Fig. 1.
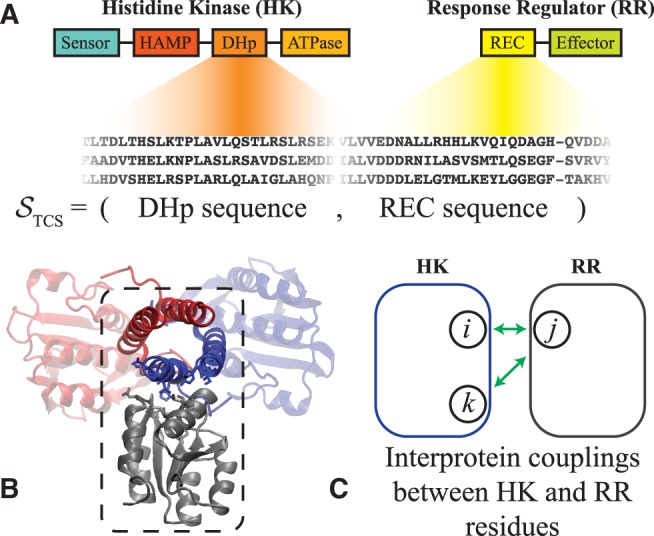
TCS domain interactions of interest. We focus only on HK proteins that have the following domain architecture from N to C terminus: sensor, HAMP, DHp, and ATPase. Likewise, we consider RR proteins that consist of a REC domain followed by an effector domain. (A) The interaction between the DHp and REC domains of the HK and RR proteins, respectively, form the TCS complex. Sequences of TCS partners are collected and stored as the concatenated sequence of the DHp and REC domains, STCS (See “Materials and Methods” section). (B) A representative structure of the HK/RR TCS complex previously predicted for the KinA/Spo0F complex in B. subtilis (Cheng et al. 2014). The HK homodimer is shown in red and blue whereas the receiver domain of the RR is shown in gray. The dashed box highlights the DHp and REC interface. This predicted complex is consistent with the experimentally determined crystal structure of HK853/RR468 of T. maritima (Casino et al. 2009) as well as another computationally predicted TCS complex (Schug et al. 2009). (C) Our proxy for signal transfer efficiency, (eq. 4), is composed of the statistical coupling parameters that describe coevolution between interprotein residues (depicted in green). Hence, naturally captures the context-dependence of mutating a residue in the HK when a residue in the RR is also mutated, or vice versa (See “Materials and Methods” section for more details).
