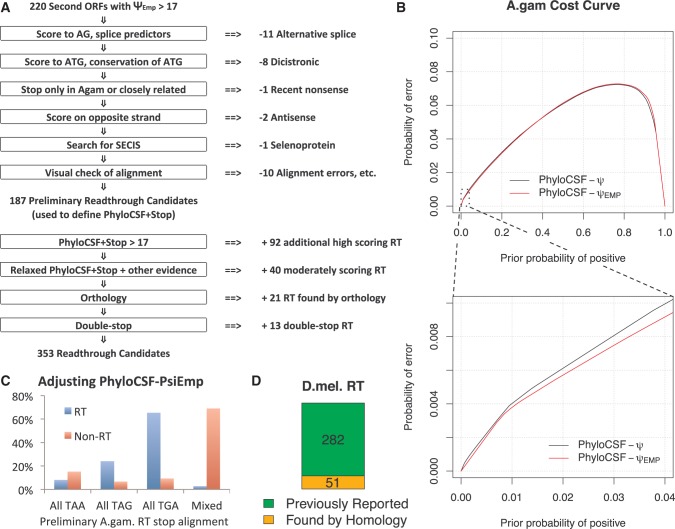
Fig. 2.
New techniques identify 353 A. gambiae and 51 additional D. melanogaster readthrough candidates. (A) Steps used to generate list of readthrough candidates in A. gambiae. Starting with 220 second ORFs having high PhyloCSF-ΨEmp score, we eliminated cases with a more plausible explanation of the protein-coding signature to yield 187 preliminary readthrough candidates. We used these to train PhyloCSF + Stop, and used that, orthology to D. melanogaster, and other evidence to find 166 additional readthrough candidates. (B) PhyloCSF-ΨEmp is an improved method for distinguishing protein-coding regions when extremely high specificity is required. Cross-validated cost curve (Drummond and Holte 2000) shows, for each prior probability that the input region is coding, the probability that the discriminator makes an error at the optimal score threshold for that prior. The performance of PhyloCSF-Ψ and of PhyloCSF-ΨEmp are similar for most values of the prior, but when the prior probability of coding is extremely low, PhyloCSF-ΨEmp makes noticeably fewer errors, for example, 7% fewer errors when the prior probability is 2%. (C) Figure shows the fraction of preliminary readthrough candidate first stop codons and other stop codons for which all aligned stop codons are TAA, TAG, TGA, or a mix. For most preliminary readthrough candidates, the first stop codon is perfectly conserved, usually TGA, whereas the majority of other annotated stop codons are not. We used this to define PhyloCSF + Stop of a second ORF by determining to which of these four categories its first stop codon belongs, and combining that evidence with its PhyloCSF-ΨEmp score. (D) For our comparative analyses, we used 333 D. melanogaster readthrough candidates consisting of 282 that had been reported in our earlier paper and 51 newly reported readthrough candidates found by homology to our A. gambiae candidates or the other D. melanogaster candidates.