ABSTRACT
The stringency of crRNA-protospacer DNA base pair matching required for effective CRISPR-Cas interference is relatively low in crenarchaeal Sulfolobus species in contrast to that required in some bacteria. To understand its biological significance we studied crRNA-protospacer interactions in Sulfolobus islandicus REY15A which carries multiple, and functionally diverse, interference complexes. A range of mismatches were introduced into a vector-borne protospacer that was identical to spacer 1 of CRISPR locus 2, with a cognate CCN PAM sequence. Two important crRNA annealing regions were identified on the 39 bp protospacer, a strong primary site centered on nucleotides 3 – 7 and a weaker secondary site at nucleotides 21 – 25. Multiple mismatches introduced into remaining protospacer regions did not seriously impair interference. Extending the study to different protospacers demonstrated that the efficacy of the secondary site was greatest for protospacers with higher G+C contents. In addition, the interference effects were assigned specifically to the type I-A dsDNA-targeting module by repeating the experiments with mutated protospacer constructs that were transformed into an S. islandicus mutant lacking type III-Bα and III-Bβ interference gene cassettes, which showed similar interference levels to those of the wild-type strain. Parallels are drawn to the involvement of 2 annealing sites for microRNAs on some eukaryal mRNAs which provide enhanced binding capacity and specificity. A biological rationale for the relatively low crRNA-protospacer base pairing stringency among the Sulfolobales is considered.
KEYWORDS: Cas, CRISPR, cRNA, protospacer, Sulfolobus, Type I-A
Introduction
Archaeal CRISPR-Cas adaptive immune systems fall into 2 major classes denoted type I and type III.1,2 The immune response involves 3 main steps, adaptation, biogenesis of crRNAs and interference. Adaptation involves incorporation of small DNA fragments from invading genetic elements into CRISPR arrays as de novo spacer-repeat units generally, but not invariably, adjacent to the leader region.3,4 Moreover, in Sulfolobus species, a single adaptation module is often cofunctional with functionally diverse interference modules.1,5 crRNAs are generated by processing of CRISPR transcripts by Cas6 cleavage within repeats. They carry most or all of the spacer sequence, depending on whether they participate in type I or type III interference, with an 8 nt repeat sequence at the 5´-end.6-9 These crRNAs assemble with Cas proteins into effector complexes and anneal to spacer-matching sequences (protospacers) on invading genetic elements, or transcripts thereof, which are then cleaved.10-12
Much of the seminal work on crenarchaeal CRISPR-Cas systems has been performed on a few model Sulfolobus species, including Sulfolobus solfataricus P2 and S. islandicus REY15A, which can host a variety of diverse viruses and for which versatile genetic systems have been developed (reviewed in13-15). These CRISPR-Cas systems tend to be exceptional in that CRISPR loci are multiple and large, and a mixture of type I-A and different type III systems are generally present.1 Although Sulfolobus CRISPR transcription and processing mechanisms have been elucidated and the modes of CRISPR-Cas adaptation and interference have been studied extensively (reviewed in 13,14), questions remain regarding the degree of crRNA-protospacer base pairing stringency required for effective interference.16-18
In initial studies on cRNA-protospacer annealing in the bacteria-specific type II CRISPR-Cas system of Streptococcus thermophilus, effective interference was inferred to require highly stringent base pairing of the crRNA-protospacer sequences.19,20 Moreover, results obtained with type I-E system of Eschericia coli also indicated the involvement of additional strict base pair matching at positions 1–5 and 7–8 of the protospacer that provides a seeding sequence for crRNA annealing21 and similar results were observed for the type I-F system of Pseudomonas aeroginosa.22 For the type I-B system of the archaeon Haloferax volcanii matching at positions 1–5, 7–10 and 13 was shown to be critical for effective interference.23 However, more recent studies on the latter type I-B, I-E and I-F systems have suggested that less perfect annealing may still be effective, depending on the sequence of the protospacer and on the spacer-specific crRNA yields.22-25
In contrast, studies on crRNA-protospacer annealing in Sulfolobus species have indicated that a much lower level of base pair matching can still produce effective interference. Experiments employing a plasmid carrying viral genes, or protospacers, that match CRISPR spacers of S. solfataricus P2 or S. islandicus REY15A demonstrated that the interference response could tolerate several base pair mismatches, 16 and similar conclusions were reached in a parallel study with modified SSV1 fusellovirus infecting S. solfataricus P2.17 The latter study was later extended to demonstrate that base pairing toward the start of the protospacer was relatively more important for strong interference.18
Nevertheless, interpretation of crRNA annealing properties in Sulfolobus species is complicated by the coexistence of multiple effector complexes which compete for similar crRNAs but exhibit different targeting mechanisms. Both S. solfataricus P2 and S. islandicus REY15A carry type I-A, type III-Bα and III-Bβ interference modules, targeting dsDNA, transcribing DNA and transcripts, respectively, 10-12,16,17,26 and the former organism also carries a type III-D module which may function similarly to the type III-Bα module.1,27 To date no experimental distinction has been made between the crRNA annealing requirements of these different effector complexes. Furthermore, there is still no credible explanation for the strong differences in base pair stringency requirements observed between Sulfolobus species and those of some bacterial species carrying type I-E and type I-F CRISPR-Cas systems.21,22,24,25
In this study, we utilize a protospacer-carrying plasmid vector system developed earlier16 to investigate more systematically the dependence of interference on the stringency of crRNA-protospacer annealing of CRISPR locus 2-spacer 1 (L2S1) of S. islandicus REY15A. Moreover, we establish that PAM-dependent type I-A interference was specifically monitored by employing a knockout mutant lacking the gene cassettes for the type III-Bα and III-Bβ interference modules.12 Introduction of a large variety of different combinations of base mutations along the protospacer reaffirmed that multiple mismatched base pairs generally produced interference, albeit often at reduced levels. In addition, evidence was found for the presence of primary (positions 3–7) and secondary (positions 21–25) annealing sites for which base pairing was important. Analysis of these 2 sites for 5 different CRISPR spacers showed that the effectiveness of secondary annealing was dependent on the G + C content of the protospacer.
Results
Identifying important crRNA-protospacer annealing sites
Constructs were prepared in plasmid pEXA2 carrying the protospacer sequence matching spacer 1 at the leader end of CRISPR locus 2, L2S1, of S. islandicus REY15A, that carried a cognate CCN PAM (Fig. 1A). Constructs containing mutated protospacers were transformed into S. islandicus and the level of CRISPR-Cas interference was estimated by comparing the numbers of transformant colonies formed on Gelrite plates with those from control samples transformed with pEXA2 lacking the protospacer using the method described earlier.10,16 High levels of transformants in the sample carrying the active protospacer were indicative of a low level, or absence, of CRISPR-Cas interference whereas few surviving transformants resulted from strong interference, such that 100% transformation efficiency (T.E.) indicates zero interference and vice-versa. The average value and standard deviation of the numbers of transformant colonies was estimated in triplicate experiments for each protospacer mutation.
Figure 1.
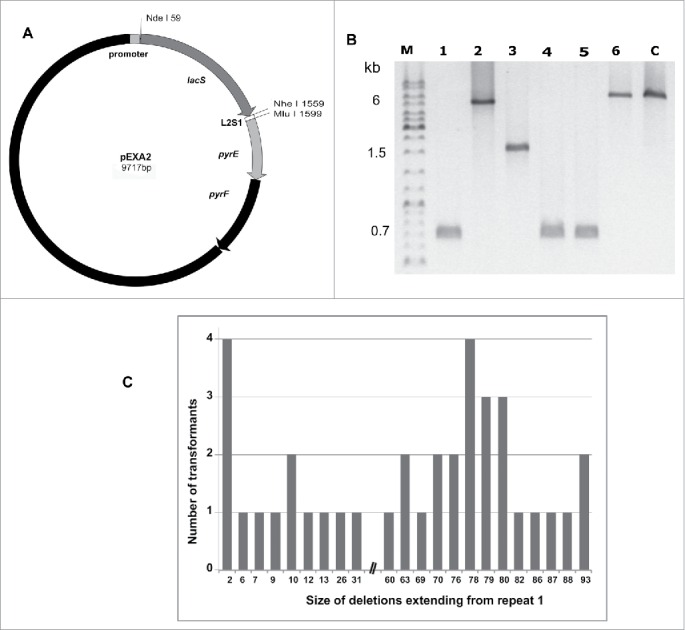
CRISPR deletions induced by plasmid-borne mutated protospacers matching spacer L2S1. (A) Protospacer L2S1 inserted into pEXA2 vector with lacS as reporter and pyrEF genes as selective markers. (B) PCR amplified products from the leader region to repeat 94 of CRISPR locus 2, from selected transformants that have survived CRISPR-Cas interference. The products were resolved in 0.8% agarose gels and carried deletions between repeats (r): lane 1: r1-r79 - no mutation; lane 2: r1-r13 - A to C (position 1); lane 3: r1-r63 – AC to CT (positions 1–2); lane 4: r1-r80 – ACA to CTG (positions 1–3); lane 5: r1-r80 - ACAC to CTGG (positions 1–4); lane 6: no deletion - ACACT to TGTGA (positions 1–5). C - control from host CRISPR locus 2. M - size markers. (C) Histogram showing the number and size distributions of all the unique CRISPR locus 2 deletions observed in surviving transformants after challenging with plasmid-borne protospacers carrying a range of mismatches. No deletions terminated between repeats 31 and 60.
In addition, many surviving transformants were tested for the presence of L2S1 by PCR amplification of the CRISPR locus 2 and a gel electrophoresis assay (Fig. 1B) and almost all constructs had retained both the protospacer and the plasmid-borne pyrEF genes that are essential for growth by undergoing deletions extending from CRISPR repeat 1 (Table S1). This resulted in the loss of both L2S1 and the capacity for CRISPR-Cas interference of the plasmid. Non identical deletions were identified in 37 transformants from different experiments; they all included L2S1 and many were large, extending to repeats 60 to 93, with no deletions terminating between repeats 31 and 60 (Fig. 1C).
Initially, 15 single-site mutations were introduced at protospacer positions 1 to 8, the location of the “seed” sequence in bacterial type I-E and I-F systems, and at sites 13, 22, 23, 24 28, 33 and 38 interspaced along the 39 bp protospacer, in order to determine potentially important recognition sites, and the sites were mutated for 3 alternative nucleotides (Table S1). Few transformants were obtained from each culture and generally no significant differences in transformation efficiency (TE) were detected relative to the control samples, for the alternative nucleotides at each position, indicative of unimpeded interference (Table S1). However, significant minor increases in T.E. (>0.2%) were observed at positions 3 (A - G), 7 (G - A), 22 (G - C) and 23 (A - G) (Table S1).
Next, we generated consecutive triplet mismatches along the protospacer, introducing primarily base transversions at each position. Only two altered triplets produced significantly altered TE values (Table S1); the value increased strongly for the mismatching triplet C4-T5-G6, and less strongly for the triplet G22-A23-C24 (Table 1). These data were consistent with the minor increases observed for the single base changes at positions 3, 7, 22 and 23 (Table 2).
Table 1.
Protospacer L2S1 carrying multiple base pair mismatches at positions 1 to 10. Averaged transformation efficiencies (TE) from triplicate experiments were normalized to values for the control plasmid constructs set to 100%. The sizes of CRISPR deletions detected in surviving transformants are shown with the repeat (r) extremities. 0 - no deletion detected. n.d. not determined. The major annealing site is centered on the shaded positions.
| Construct | 1A | 2C | 3A | 4C | 5T | 6G | 7G | 8G | 9C | 10G | TE (%) | CRISPR deletion |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | C | T | < 0.1 | r1-r63 | ||||||||
| 2 | C | A | G | < 0.1 | r1-r63 | |||||||
| 3 | C | T | G | 0.1 | r1-r80 | |||||||
| 4 | C | T | G | T | 0.2 | r1-r70 | ||||||
| 5 | C | T | G | G | 1.1 | r1-r80 | ||||||
| 6 | C | T | G | A | 13 | 0 | ||||||
| 7 | T | G | T | < 0.1 | n.d. | |||||||
| 8 | T | G | T | G | 12 | 0 | ||||||
| 9 | C | T | G | A | C | 17 | 0 | |||||
| 10 | T | G | T | G | A | 24 | 0 | |||||
| 11 | G | T | G | A | 9 | 0 | ||||||
| 12 | G | T | G | A | C | 17 | 0 | |||||
| 13 | T | G | A | C | C | 79 | 0 | |||||
| 14 | G | A | C | 10 | 0 | |||||||
| 15 | A | C | C | C | 0.1 | n.d. | ||||||
| 16 | C | C | C | G | C | < 0.1 | n.d. | |||||
| 17 | C | C | G | < 0.1 | n.d. |
Table 2.
Protospacer L2S1 carrying multiple base pair mismatches at positions 19 to 27. Averaged transformation efficiencies (TE) from triplicate experiments were normalized to values for control plasmid constructs set to 100%. The sizes of CRISPR deletions in surviving transformants are given with repeat (r) extremities. n.d. - not determined. The minor annealing site is centered on the shaded positions.
| Construct | 19A | 20A | 21C | 22G | 23A | 24C | 25A | 26G | 27G | TE (%) | CRISPR deletion |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | T | T | G | 0.1 | n.d. | ||||||
| 2 | T | T | G | C | T | 1 | r1-r7 | ||||
| 3 | T | G | C | T | G | T | C | 10 | n.d. | ||
| 4 | G | C | T | G | T | 6 | n.d. | ||||
| 5 | C | T | G | T | 1.1 | r1-r26 | |||||
| 6 | C | T | G | 1 | r1-r7 | ||||||
| 7 | T | G | T | C | 1.2 | n.d. | |||||
| 8 | C | T | < 0.1 | n.d. | |||||||
| 9 | T | G | 0 | n.d. | |||||||
| 10 | C | G | 0 | n.d. | |||||||
| 11 | T | C | C | < 0.1 | n.d. |
Consecutive quintuplet base pair mismatches were then generated along the protospacer, most with base transversions (Table S1). Strongly increased TE values were observed for mutated quintuplets 1 – 5 and 2 – 6 (17 – 24%) and a very strong increase occurred for positions 3 – 7 (79%) (Table 1). No other significant differences from the control were observed except for the quintuplet at positions 21 – 25, where a moderately increased TE value (6%) was detected (Table 2). Finally, mismatches were introduced at positions 11 – 20, 20 – 26, 21 – 30 and 31 – 39; the former 3 produced moderately increased TE values while the latter was indistinguishable from the control (Table S1).
A summary of the data for the site at the start of the protospacer showed that whereas mismatches at positions 1, 1+2 had no effect on TE levels, with minor increases for mutations at positions 1+2+3. Results obtained for positions 1 to 4 were variable and dependent on the mismatching nucleotides. Overall the data support that positions 3 to 7 are critical for the annealing process (Table 1).
At the second site, beyond the center of the protospacer, alignment of the corresponding sets of data show that transformation efficiencies increased significantly for mutated positions 20 – 26 (10%) and 21 – 25 (6%), but were low for all partially overlapping sequences tested including 19 – 23 and 23 – 26 (1–1.2%) and all less strong than for positions 3 to 7 (Table 2). In summary, the results are consistent with the the presence of major and minor crRNA annealing sites centered on positions 3 – 7 and 21 – 25, respectively, that are important for effective interference.
Which type of effector complex targets the plasmid protospacer?
S. islandicus encodes 3 CRISPR-Cas interference complexes which compete for similar crRNAs, type I-A, type III-Bα and III-Bβ which cleave dsDNA, transcripts and transcribing DNA, and RNA transcripts, respectively (Fig. 2). Although this, and the earlier experiments, were designed to test for type I-A-directed DNA interference, we could not eliminate the possibility that transcriptional read-through from distant promoters had occurred on either strand of the protospacer, such that type III interference was also activated. Therefore, we repeated the experiments in an S. islandicus mutant lacking type III-Bα + III-Bβ interference gene cassettes.12 Protospacer constructs were prepared with a cognate CCN PAM and mismatches were introduced in sequence regions 3 – 7 and 21 – 25. No significant differences were observed in the levels of interference observed for the mutant and wild-type strains (Fig. 3) and it was concluded, therefore, that all interference effects observed derived primarily from type I-A effector complexes.
Figure 2.
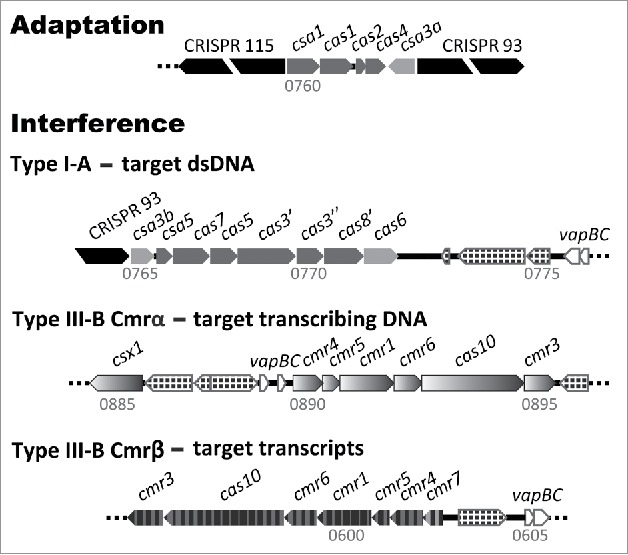
Overview of cas gene cassettes of the single adaptation and 3 interference CRISPR-Cas modules of S. islandicus REY15A. CRISPR loci 1 (115 repeat-spacer units) and 2 (93 units) are indicated and the genomic locations are given. Antitoxin-toxin vapBC gene pairs are shown together with mobile element-related genes (chequered).
Figure 3.
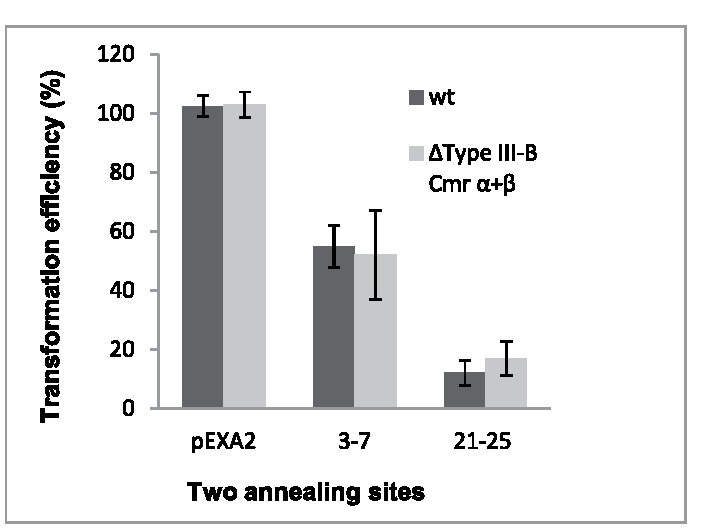
Averaged transformation efficiencies of constructs of the L2S1 protospacer carrying quintuplet mismatches at positions 3 – 7 or 21 – 25. Each construct was transformed into the hosts: S. islandicus E233 strain (black) and S. islandicus E233 ΔIII-B Cmr-α + Cmr-β interference modules (gray). Transformation efficiency of the control plasmid is set at 100%. Error bars are shown for triplet experiments for each construct.
PAM sequence recognition during interference
Although the dinucleotide PAM sequence required for CRISPR-Cas adaptation in Sulfolobus species appears to be highly conserved, and is CCN for most, but not all, Sulfolobus type I-A subfamilies, 3,16 earlier studies suggested that some sequence redundancy in the dinucleotide was tolerated during interference.16 In addition, studies on haloarchaeal type I-B interference revealed that a range of triplet PAM sequences ACT, TAA, TAT, TAG, TTC, and CAC were active.28 Therefore, using the same plasmid-borne protospacer assay, we examined systematically the effects of all the alternative dinucleotide CCN PAM sequences in wild-type S. islandicus and in the deletion mutant lacking type III-B Cmr-α + Cmr-β interference gene cassettes.12 The PAM analysis data demonstrate that CCA is the most effective PAM. It is about 5-fold more effective than TCA and CTA which, in turn, are about 10-fold better than the remainder of the PAMs (Fig. 4; Table S2).
Figure 4.
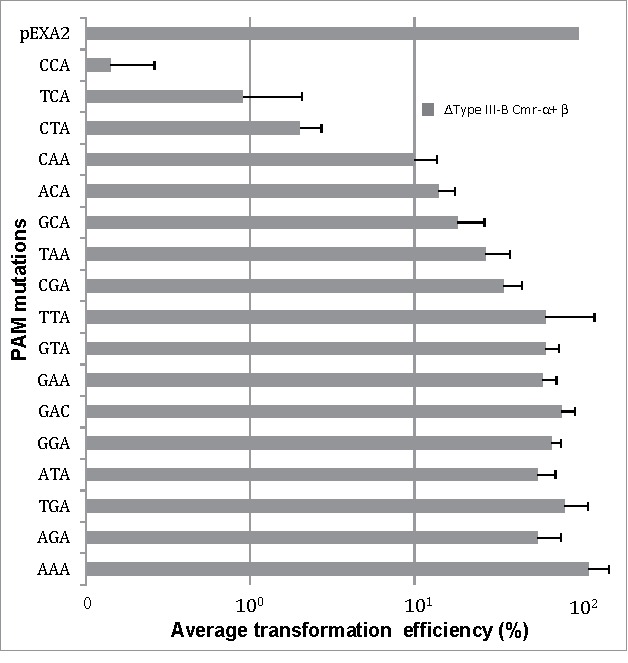
Averaged transformation efficiencies of the S. islandicus ΔIII-Bα and IIIB-β deletion mutant after challenging with plasmid-borne L2S1 protospacers carrying all possible dinucleotide modifications of the CCN PAM sequence. Transformation efficiency of the non-target plasmid is set at 100% and error bars derive from triplet experiments for each construct. Transformation efficiency and standard deviation values for the wild-type and mutated strains are listed in Table S2.
Are the 2 annealing regions conserved in other protospacers?
The variability in sequence, base composition and size of CRISPR spacers could influence the preferred annealing sites and, therefore, the experiments were repeated with protospacers matching 4 additional CRISPR spacers with different G+C contents: locus 1-spacer 58 (61.5% GC), locus 2-spacer 55 (60.5% GC), locus 2-spacer 45 (49% GC) and locus 1-spacer 61 (25.6% GC). Transversion mutations were generated in the quintipulet positions 3 – 7, 11 – 15, 16 – 20, 21 – 25 and 31 – 35 of the plasmid-borne protospacers to test for positional conservation of the observed annealing sites. The average transformation efficiencies observed are summarised in Table 3 and they correlate approximately with the results obtained for L2S1, with interference being significantly reduced only for the constructs modified at positions 3 – 7 and 21 – 25. However, the effectiveness of both sites was dependent on the overall G+C content of the protospacer. The largest effects were observed for the 2 spacers with 61.5% G+C decreasing through 60.5% and with reduced but significant effects at 49% and 25.6% G+C (Table 3).
Table 3.
Normalized average transformation efficiencies obtained with plasmid-borne protospacers matching 5 different spacers of CRISPR loci 1 and 2. The constructs carry quintipulet mismatches with transversions distributed along the protospacer. Experiments were performed in triplicate. Results for the major and minor annealing sites are shaded.
| CRISPR spacer – TE% values |
|||||
|---|---|---|---|---|---|
| Mutated quintuplet | L2-S1 (61.5% GC) | L1-S58 (61.5% GC) | L2-S55 (60.5% GC) | L2-S45 (49% GC) | L1-S61 (25.6% GC) |
| 3 – 7 | 79 (±7.1) | 87.3 (±19.9) | 66.3 (± 11.2) | 41.5 (± 11.5) | 25 (± 2.5) |
| 11 – 15 | 0.031 (±0.41) | 0.17 (±0.12) | 0. 1 (± 0. 1) | 0.2 ( ± 0.1) | 0.16 (± 0.14) |
| 16 – 20 | 0.002 (±0.002) | 0.16 (±0.05) | 0.1 (± 0.04) | 0.01 (± 0.01) | 0.04 (± 0.01) |
| 21 – 25 | 6 (±4.2) | 7.4 (± 5.7) | 3.4 (± 1.7) | 1.05 (± 0.05) | 0.8 (± 0.1) |
| 31 – 35 | 0.023 (±0.032) | 0.6 (±0.2) | 0.7 (±0.3) | 0. 3 (± 0.03) | 0.44 (± 0.07) |
Discussion
Earlier studies on the stringency of crRNA-protospacer base pairing in different Sulfolobus species, utilizing plasmid- and fusellovirus SSV1-borne protospacers that were identical to host CRISPR spacers, demonstrated that effective interference could occur despite the presence of multiple base pair mismatches.16,17 Later the viral study was extended to demonstrate that disruption of the base pairing near the start of the protospacer was particularly effective in impairing interference.18 These results are summarised in Table 4 and, overall, they differ strongly from data obtained for some bacterial CRISPR-Cas systems, including the type II system of S. thermophilus19,20 and bacterial type I-E and I-F systems21,22,24,25 which require high base pairing stringency for effective interference.
Table 4.
Summary of literature experiments investigating the stringency of crRNA-protospacer base pairing required for effective interference for selected CRISPR spacers of Sulfolobus species. TE - transformation efficiency; pfu - plaque forming units.
| Organism/CRISPR spacer/PAM | Vector | Mutated protospacer positions | TE (%) |
|---|---|---|---|
| S. solfataricus P216 LA-S28 (TCN) | plasmid | 1, 19, 38, 1+38, 10+19, 35−38, 29−38 | <2 |
| LD-S29 (CCN) | plasmid | 24, 18+24, 18+21+24 | <2 |
| S. islandicus16 L2-S45 (CCN) | plasmid | 1, 19, 38, 34–37, 29−32+35−38 | <2 |
| pfu (%) | |||
| S. solfataricus P217 LA-S53 (TCN) | SSV1 virus | 2+5−6 | 85 |
| 2+5−6+18+32+34+36 | 20 | ||
| S. solfataricus P2 18 LA-S53 (TCN) | SSV1 virus | 1–6, 1–8, 1–10, 2+5−6 | 75, 87, 80, 78 |
| 2+5−6+32−35+37 | 29 | ||
| 32+34, 32−35+37 | 0, 4 | ||
| 29−35+37, 27−35+37 | 34, 43 | ||
| 25+27−35+37 | 65 | ||
| 24−25+27−35+37 | 30 | ||
| 23−25+27−35+37 | 54 | ||
| 20−25+27−35+37 | 90 | ||
In the present work, by introducing diverse single and multiple nucleotide mutations along a protospacer, we could demonstrate that effective interference could tolerate mismatches all along the protospacer but that 2 regions are particularly critical for interference, a major site centered on positions 3 to 7 and a minor site at positions 21 – 25, for different protospacers. These findings are consistent with, and significantly extend, the earlier Sulfolobus results (Table 4). However, Sulfolobus species generally carry multiple type I-A and type III CRISPR-Cas interference modules and experiments, to date, have not established which type of crRNA-directed interference was being monitored. Therefore, we employed an S. islandicus deletion mutant lacking gene cassettes for the type III-Bα and III-Bβ effector complexes which target transcribing DNA and transcripts, respectively.12 The results demonstrated that only type I-A-directed DNA interference was being measured.
The importance of a single primary seeding site for crRNA-protospacer annealing was first demonstrated for the type I-E system of E. coli21 and involved protospacer positions 1 – 5 and 7 – 8, and more recently similar seed sequences have been observed for the type I-E and I-F systems of Pseudomonas aeruginosa22 and Pectobacterium atrosepticum29 and, with some variation, for the type I-B system of Haloferax volcanii.23 A rationale for the non involvement of position 6 in the type I-E system was later provided by the crystal structure of the E. coli type I-E effector complex in which every sixth nucleotide (positions 6, 12, 18 etc.) in the crRNA-DNA duplex was stabilised by Cas proteins and unavailable for base pairing.30 The comparable annealing site in Sulfolobus is smaller, and less stringent, with single mutations of each of the individual nucleotides being tolerated (Table S1). Moreover, the structure of the crenarchaeal type I-A interference complex differs substantially from that of the bacterial type I-E complex; they share a Cas7 oligomeric backbone and a base structure of Cas5 and Cas8 but, in contrast to the bacterial complex, the type I-A complex carries Cas3′ and Cas3” but no Cas6.21,31-33 Therefore, the structural details of the crRNA-DNA-protein interactions are likely to differ significantly.
A possible insight into the different crRNA annealing properties in Sulfolobus derives from a study of type I-E interference in E. coli which demonstrated that 2 distinct crRNA-DNA mechanisms can operate34; the one described above that involves stringent base pairing that is essential for interference, and another with less stringent base pairing that may facilitate priming of CRISPR-Cas adaptation, a process that has not been demonstrated unambiguously to occur in Sulfolobus species.13,35 Possibly, Sulfolobus type I-A interference uses a more primitive system wherein the minor annealing site increases the overall binding specificity.
The operation of less specific interference could also explain the less strict CCN PAM recognition for interference, 16,36 in contrast to the strict requirement for the CCN PAM during adaptation.4 About 5-fold weaker effects were observed with TCA and CTA, consistent with earlier work, 16 and even weaker effects were observed with GCN, CAN, ACN, suggesting that CN and NC pairs can elicit an interference response.
The finding of 2 annealing sites also suggests a structural similarity to the mode of action of some microRNAs (miRNA). These small 20 – 25 nt RNAs regulate gene expression in animals and plants by binding to UTRs of mRNAs.37 Moreover, they carry a “seed” region at 5′-positions 2 – 7 which constitutes the primary annealing site to mRNAs but many miRNAs also exhibit a secondary binding site toward the 3′-end (positions 13 – 16) which may have a compensatory role when the 5´-seed interaction is weaker and/or generate enhanced specificity for certain mRNA sites.38-40
Most of the tested transformants that survived interference were found to have undergone deletions from CRISPR locus 2, that included L2S1, and they tended to be large, often covering much of the array. Such deletions were observed earlier when a plasmid-borne protospacer was constructed with pyrE/F genes that were essential for host cell growth. The deletions were inferred to arise from rare, random, recombination events between repeats, 16,41 which is consistent with the finding that recombination can occur between relatively short sequences in Sulfolobus.42 An alternative explanation would be a reverse adaptation reaction involving Cas1 but then one would expect a high proportion of single L2S1 deletions, as was observed in an earlier S. islandicus experiment.16
The CRISPR-Cas systems of the Sulfolobales are exceptional in that CRISPR loci tend to carry hundreds of spacers, including multiple spacers with significant sequence matches to specific viruses or conjugative plasmids.1,5 Paradoxically, some Sulfolobus species are also excellent hosts for viral propagation in the laboratory. Therefore, it has been suggested that this extensive spacer redundancy, and partially overlapping interference capabilities, could reflect that the interference is relatively inefficient and primarily directed to restricting viral infections to manageable levels for the host.5,13 This would have the considerable advantage that the CRISPR-Cas adaptation, which coincides with strong growth retardation and extensive cell death, could generally be avoided,.35,43,44
Materials and methods
Archaeal and bacterial strains, media and growth conditions
S. islandicus E233S is a uracil-auxotroph of S. islandicus with the complete pyrE/pyrF and lacS genes deleted.45 Sulfolobus SCV medium was used and 0.2% sucrose, 0.2% (w/v) vitamin free casamino acids, and a mixed vitamin solution, were added and the final pH was adjusted to 3.5 using 1 M H2SO4.46 Sulfolobus strains, or colonies, were inoculated into test tubes containing 6 mL SCV and grown at 78 °C with shaking at 150 rpm. Sulfolobus colonies were developed on 0.7% Gelrite plates. Two-layer plating was used where diluted liquid cultures were mixed with the same medium containing 0.4% Gelrite and plated on an upper layer. Strains or transformants to be selected via uracil drop out selection were cultured in SCV medium, whereas pyrEF-minus mutants were cultured in SCV with added uracil (20 mg/mL).10, 16
E. coli DH5α and pEXA2 were employed as host and cloning vector, respectively. Transformants were cultured in Luria–Bertani (LB) medium supplemented with 100 mg/ml ampicillin and cultured at 37 °C. Standard cloning methods were used for DNA manipulation.47 Restriction enzymes were from New England Biolabs (Hitchin, UK) or Fermentas (Waltham, MA). Plasmid DNA was isolated from E. coli and Sulfolobus cells using QIAprep Spin Miniprep kits (QIAGen Westberg, Germany). Total DNA was prepared using QIAGen DNeasy kits (Hilden, Germany). DNA oligonucleotides used for PCR amplification were synthesized at TAG (Copenhagen, Denmark) and DNA was sequenced by Eurofins MWG (Ebersberg, Germany).
Plasmid constructs
All plasmid constructs were derived from the shuttle vector pEXA2 carrying pyrEF genes.16 The lacS gene cassette was amplified from S. islandicus E233 by PCR using specific primers.45 NheI and NdeI restriction sites were incorporated into the primers to facilitate cloning. The DNA polymerase used for producing PCR fragments was purified with the QIAGen PCR purification kit.
Spacer sequences SisL2S1, SisL2S45, L2S55, L1S58 and SisL1S61 were used to design forward and reverse primers carrying NheI and MluI restriction sites with overlaps. The primer sets were annealed by heating for 5 min at 95 °C and cooling to room temperature on a Thermo-block (Eppendorf, Hamburg, Germany) and maintaining at 4 °C for 15 min. Ligation was performed to insert annealed protospacer fragments into purified digested pEXA2 vector for 3 h at 22 °C. The ligation mixture was transformed into E. coli DH5α competent cells using heat shock and harvested on Luria-Bertani (LB) plates supplemented with 100 mg/ml ampicillin and incubated at 37 °C for 12 – 16 h. Positive colonies were detected by colony PCR and cultured in LB medium supplemented with 100 mg/ml ampicillin, incubating at 37 °C for 10 – 14 h.47 Plasmids were purified using Miniprep plasmid purification kits (QIAGen) and precipitated with ethanol.
Transformation procedures
S. islandicus transformation was generally performed by electroporation48 and all procedures were executed at room temperature and 950 µL preheated high salt medium (pH 5 to 6) was added immediately to electroporated cells with further incubation at 75 °C for 30 min without shaking. Different dilutions of transformation mixtures were added to 8 mL of 2 x SCV medium + 0.4% Gelrite or phytagel and plated. After polymerizing the upper layer, plates were incubated in tightly closed plastic boxes at 78 °C for 5 to 8 days.10,16
The two-layer cultivation method was adopted to plate Sulfolobus transformants because non-transformants also form small colonies on selective plates on direct plating. This probably occurs because pyrimidine compounds are released from lysed cells and can support growth. Plating electroporated cells in the top Gelrite layer protects cells from lysis and only true transformants form colonies on selective plates.10,16 Typically, shuttle plasmid-transformed cells appeared as single colonies after 5 – 7 days. Furthermore, the β-glycosidase activity encoded by lacS in Sulfolobus colonies (lacS activity) was employed to reveal real transformants carrying plasmids and was detected by X-gal (5-bromo-4-chloro-3-indolyl-b-Dgalactopyranoside) staining. 2 mL X-gal (2 mg/mL) was added onto plates with transformants carrying the lacS gene a a reporter. Plates were rotated gently to distribute X-gal on the surface and incubated at 78 °C. The resulting blue color developed overnight. lacS activity was measured in liquid culture by adding X-gal to a final concentration of 2 mg/mL and incubating at 78 °C for 2 h before estimating color development.49
After selecting blue colonies that appeared in the upper Gelrite layer after 5 to 7 days, relative transformation efficiency values were calculated from the cfu per mg DNA of the construct divided by the cfu per mg DNA of the positive control plasmid, in triplicate replicates, and the values were averaged and standard deviations estimated.10,16 Relative T.E. values are presented as percentages where 100% T.E. corresponds to zero CRISPR-Cas interference and 0% T.E. denotes total interference.
Characterization of transformants and gene deletion mutants
Transformant colonies were picked from plates with a tip carrying a broad hole and transferred to the surface of another SCV plate and stored at 78 °C for 3 – 5 days. The large transformant colonies were released into SCV medium (6 mL) in a test tube and incubated at 78 °C for 3 – 7 days. After culturing, DNA was extracted from each sample and subjected to PCR amplification of CRISPR locus 2 and sequencing to check for the occurrence, and size, of CRISPR deletions. Once a desired mutant/strain was identified, each single colony was purified by transferring to a fresh SCV plate and then to SCV medium 3 successive times.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplementary Material
Acknowledgments
Members of the Danish Archaea Centre are thanked for their support and insightful discussions. The research was supported by the Danish Natural Science Research Council and Marzieh Mousaei received a PhD grant from the Ministry of Science, Research and Technology of Iran.
ORCID
Marzieh Mousaei http://orcid.org/0000-0002-3488-2609
Qunxin She http://orcid.org/0000-0002-4448-6669
References
- 1.Vestergaard G, Garrett RA, Shah SA. CRISPR adaptive immune systems of Archaea. RNA Biol 2014; 11:157-68; PMID:24531374; http://dx.doi.org/ 10.4161/rna.27990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH, et al.. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol 2015; 13:722-36; PMID:26411297; http://dx.doi.org/ 10.1038/nrmicro3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lillestøl RK, Shah SA, Brügger K, Redder P, Phan H, Christiansen J, Garrett RA. CRISPR families of the crenarchaeal genus Sulfolobus: bidirectional transcription and dynamic properties. Mol Microbiol 2009; 72:259-72; PMID:19239620; doi.org/ 10.1111/j.1365-2958.2009.06641.x [DOI] [PubMed] [Google Scholar]
- 4.Erdmann S, Garrett RA. Selective and hyperactive uptake of foreign DNA by adaptive immune systems of an archaeon via two distinct mechanisms. Mol Microbiol 2012; 85:1044-56; PMID:22834906; http://dx.doi.org/ 10.1111/j.1365-2958.2012.08171.x Corrigendum: Mol Microbiol 86: 757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shah SA, Garrett RA. CRISPR/Cas and Cmr modules, mobility and evolution of adaptive immune systems. Res Microbiol 2011; 162:27-38; PMID:20863886; http://dx.doi.org/ 10.1016/j.resmic.2010.09.001 [DOI] [PubMed] [Google Scholar]
- 6.Tang TH, Bachellerie JP, Rozhdestvensky T, Bortolin ML, Huber H, Drungowski M, Elge T, Brosius J, Hüttenhofer A. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc Natl Acad Sci USA 2002; 99:7536-41; PMID:12032318; http://dx.doi.org/ 10.1111/j.1365-2958.2004.04428.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 2008; 321:960-64; PMID:18703739; http://dx.doi.org/ 10.1126/science.1159689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 2009; 139:945-56; PMID:19945378; http://dx.doi.org/ 10.1016/j.cell.2009.07.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. Sequence and structure-specific RNA processing by a CRISPR endonuclease. Science 2010; 10:1355-58; PMID:20829488; http://dx.doi.org/ 10.1126/science.1192272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng L, Garrett RA, Shah SA, Peng X, She Q. A novel interference mechanism by a type IIIB CRISPR-Cmr module in Sulfolobus. Mol Microbiol 2013; 87:1088-99; PMID:23320564; http://dx.doi.org/ 10.1111/mmi.12152 [DOI] [PubMed] [Google Scholar]
- 11.Zebec Z, Manica A, Zhang J, White MF, Schleper C. CRISPR-mediated targeted mRNA degradation in the archaeon Sulfolobus solfataricus. Nucleic Acids Res 2014; 42:5280-88; PMID:24603867; http://dx.doi.org/ 10.1093/nar/gku161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peng W, Feng M, Feng X, Liang YX, She Q. An archaeal CRISPR type III-B system exhibiting distinctive RNA targeting features and mediating dual RNA and DNA interference. Nucleic Acids Res 2015; 43:406-17; PMID:25505143; http://dx.doi.org/ 10.1093/nar/gku1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garrett RA, Shah SA, Erdmann S, Liu G, Mousaei M, Leon-Sobrino C, Peng W, Gudbergsdottir S, Deng L, Vestergaard G, et al.. CRISPR-Cas adaptive immune systems of the Sulfolobales: Unravelling their complexity and diversity. Life 2015; 5:783-817; PMID:25764276; http://dx.doi.org/ 10.3390/life5010783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manica A, Schleper C. CRISPR-mediated defense mechanisms in the hyperthermophilic archaeal genus Sulfolobus. RNA Biol 2013; 10:671-78; PMID:23535277; http://dx.doi.org/ 10.4161/rna.24154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang C, Tian B, Li S, Ao X, Dalgaard K, Göcke S, Liang Y, She Q. Genetic manipulation in Sulfolobus islandicus and functional analysis of DNA repair genes. Biochem Soc Trans 2013; 41:405-10; PMID:23356319; http://dx.doi.org/ 10.1042/BST20120285 [DOI] [PubMed] [Google Scholar]
- 16.Gudbergsdottir S, Deng L, Chen Z, Jensen JV, Jensen LR, She Q, Garrett RA. Dynamic properties of the Sulfolobus CRISPR/Cas and CRISPR/Cmr systems when challenged with vector-borne viral and plasmid genes and protospacers. Mol Microbiol 2011; 79:35-49; PMID:21166892; http://dx.doi.org/ 10.1111/j.1365-2958.2010.07452.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manica A, Zebec Z, Teichmann D, Schleper C. In vivo activity of CRISPR-mediated virus defence in a hyperthermophilic archaeon. Mol Microbiol 2011; 80:481-91; PMID:21385233; http://dx.doi.org/ 10.1111/j.1365-2958.2011.07586.x [DOI] [PubMed] [Google Scholar]
- 18.Manica A, Zebec Z, Steinkellner J, Schleper C. Unexpectedly broad target recognition of the CRISPR-mediated virus defence system in the archaeon Sulfolobus solfataricus. Nucleic Acids Res 2013; 41:10509-17; PMID:24021627; http://dx.doi.org/ 10.1093/nar/gkt767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007; 315:1709-12; PMID:17379808; http://dx.doi.org/ 10.1126/science.1138140 [DOI] [PubMed] [Google Scholar]
- 20.Deveau H, Barrangou R, Garneau JE, Labonté J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol 2008; 190:1390-1400; PMID:18065545; http://dx.doi.org/21646539 10.1128/JB.01412-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Semenova E, Jore MM, Datsenko KA, Semenova A, Westra ER, Wanner B, van der Oost J, Brouns SJ, Severinov K. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci USA 2011; 108:10098-103; PMID:21646539; http://dx.doi.org/ 10.1073/pnas.1104144108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cady KC, Bondy-Denomy J, Heussler GE, Davidson AR, O'Toole GA. The CRISPR/Cas adaptive immune system of Pseudomonas aeruginosa mediates resistance to naturally occurring and engineered phages. J Bacteriol 2012; 194:5728-38; PMID:22885297; http://dx.doi.org/ 10.1128/JB.01184-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maier LK, Lange SJ, Stoll B, Haas KA, Fischer S, Fischer E, Duchardt-Ferner E, Wöhnert J, Backofen R, Marchfelder A. Essential requirements for the detection and degradation of invaders by the Haloferax volcanii CRISPR/Cas system I-B. RNA Biol 2013; 10:865-74; PMID:23594992; http://dx.doi.org/ 10.4161/rna.24282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xue C, Seetharam AS, Musharova O, Severinov K, Brouns SJJ, Severin AJ, Sashital DG. CRISPR interference and priming varies with individual spacer sequences. Nucleic Acids Res 2015; 43:10831-47; PMID:24711427:26586800; http://dx.doi.org/ 10.1093/nar/gkv1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fineran PC, Gerritzen MJH, Suarez-Diez M, Künne T, Boekhurst J, van Hijum SAFT, Staals RHJ, Brouns SJJ. Degenerate target sites mediate rapid primed CRISPR adaptation. Proc Natl Acad Sci USA 2014; 111:1629-38; PMID; http://dx.doi.org/ 10.1073/pnas.1400071111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J, Rouillon C, Kerou M, Reeks J, Brügger K, Graham SJ, Reimann J, Cannone G, Liu H, Albers SV, et al.. Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol Cell 2012; 45:303-13; PMID:22227115; http://dx.doi.org/ 10.1016/j.molcel.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang J, Graham S, Tello A, Liu H, White MF. Multiple nucleic acid cleavage modes in divergent type III CRISPR systems. Nucleic Acids Res 2016; 44:1789-99; PMID:26801642; http://dx.doi.org/ 10.1093/nar/gkw020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fischer S, Maier LK, Stoll B, Brendel J, Fischer E, Pfeiffer F, Dyall-Smith M, Marchfelder A. An archaeal immune system can detect multiple protospacer adjacent motifs (PAMs) to target invader DNA. J Biol Chem 2012; 287:33351-63; PMID:22767603; http://dx.doi.org/ 10.1074/jbc.M112.377002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vercoe RB, Chang JT, Dy RL, Taylor C, Gristwood T, Clulow JS, Richter C, Przybilski R, Pitman AR, Fineran PC. Cytotoxic chromosomal targeting by CRISPR/Cas systems can reshape bacterial genomes and expel or remodel pathogenicity islands. PloS Genet 2013; 9:e1003454; PMID ; http://dx.doi.org/ 10.1371/journal.pgen.1003454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wiedenheft B, van Duijn E, Bultema JB, Waghmare SP, Zhou K, Barendregt A, Westphal W, Heck AJ, Boekema EJ, Dickman MJ, et al.. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci, USA 2011; 108:10092-97; PMID:21536913; http://dx.doi.org/ 10.1073/pnas.1102716108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lintner NG, Kerou M, Brumfield S.K, Graham S, Liu H, Naismith JH, Sdano M, Peng N, She Q, Copie V, et al.. Structural and functional characterization of an archaeal CASCADE complex for CRISPR-mediated viral defense. J Biol Chem 2011; 286:21643-56; PMID:21507944; http://dx.doi.org/2512348 10.1074/jbc.M111.238485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plagens A, Tripp V, Daume M, Sharma K, Klingl A, Hrle A, Conti E, Urlaub H, Randau L. In vitro assembly and activity of an archaeal CRISPR-Cas type I-A Cascade interference complex. Nucleic Acids Res 2014; 42:5125-38; PMID:24500198; http://dx.doi.org/2512348 10.1093/nar/gku120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mulepati S, Héroux A, Bailey S. Crystal structure of a CRISPR RNA-guided surveillance complex bound to a ssDNA target. Science 2014; 345:1479-84; PMID:2512348; http://dx.doi.org/ 10.1126/science.1256996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blosser TR, Loeff L, Westra ER, Vlot M, Künne T, Sobota M, Dekker C, Brouns SJ, Joo C. Two distinct DNA binding modes guide dual roles of a CRISPR-Cas protein complex. Mol Cell 2015; 58:60-70; PMID:25752578; http://dx.doi.org/ 10.1016/j.molcel.2015.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu G, She Q, Garrett RA. Diverse CRISPR-Cas responses and dramatic cellular DNA changes and cell death in pKEF9-conjugated Sulfolobus species. Nucleic Acids Res 2016; 44:4233-42; PMID:27098036; http://dx.doi.org/ 10.1093/nar/gkw286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shah SA, Erdmann S, Mojica FJ, Garrett RA. Protospacer recognition motifs: mixed identities and functional diversity. RNA Biol. 2013; 10:891-99; PMID:23403393; http://dx.doi.org/ 10.4161/rna.23764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009; 136:215-33; PMID:19167326; http://dx.doi.org/ 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol 2005; 3:e85; PMID:15723116; http://dx.doi.org/ 10.1371/journal.pbio.0030085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chorn G, Zhao L, Sachs AB, Flanagan WM, Lim LP. Persistence of seed-based activity following segmentation of a microRNA guide strand. RNA 2010; 16:2336-40; PMID:20971811; http://dx.doi.org/ 10.1261/rna.2296210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Künne T, Swarts DC, Brouns SJJ. Planting the seed: target recognition of short guide RNAs. Trends Microbiol 2014; 22:74-83; PMID:24440013; http://dx.doi.org/ 10.1016/j.tim.2013 [DOI] [PubMed] [Google Scholar]
- 41.Deng L, Kenchappa CS, Peng X, She Q, Garrett RA. Modulation of CRISPR locus transcription by the repeat binding protein Cbp1 in Sulfolobus. Nucleic Acids Res 2012; 40:2470-80; PMID:22139923; http://dx.doi.org/ 10.1093/nar/gkr1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grogan DW. Homologous recombination in Sulfolobus acidocaldarius: Genetic assays and functional properties. Biochem Soc Trans 2009; 37:88-91; PMID:19143608; http://dx.doi.org/ 10.1042/BST0370088 [DOI] [PubMed] [Google Scholar]
- 43.Erdmann S, Le Moine Bauer S, Garrett RA. Inter-viral conflicts that exploit host CRISPR immune systems of Sulfolobus. Mol Microbiol 2014; 91:900-17; PMID:24433295; http://dx.doi.org/ 10.1111/mmi.12503 [DOI] [PubMed] [Google Scholar]
- 44.Leon-Sobrino C, Kot WP, Garrett RA. Transcriptome changes in STSV2-infected Sulfolobus islandicus REY15A undergoing continuous de novo CRISPR spacer acquisition. Mol. Microbiol 2016; 99:719-28; PMID:26514343; http://dx.doi.org/ 10.1111/mmi.13263 [DOI] [PubMed] [Google Scholar]
- 45.Deng L, Zhu H, Chen Z, Liang YX, She Q. Unmarked gene deletion and host-vector system for the hyperthermophilic crenarchaeon Sulfolobus islandicus. Extremophiles 2009; 13:735-46; PMID:19513584; http://dx.doi.org/ 10.1007/s00792-009-0254-2 [DOI] [PubMed] [Google Scholar]
- 46.Zillig W, Kletzin A, Schleper C, Holz I, Janekovic D, Hain J, Lanzendörfer M, Kristjansson JK. Screening for Sulfolobales, their plasmids and their viruses in Icelandic solfataras. Syst. Appl Microbiol 1993; 16:609-28; http://dx.doi.org/ 10.1016/S0723-2020(11)80333-4 [DOI] [Google Scholar]
- 47.Sambrook J, Russell D. Molecular Cloning: a Laboratory Manual, 3rd edn. Cold Spring Harbor 2001, NY: Cold Spring Harbor Laboratory [Google Scholar]
- 48.Schleper C, Kubo K, Zillig W. The particle SSV1 from the extremely thermophilic archaeon Sulfolobus is a virus: demonstration of infectivity and of transfection with viral DNA. Proc Natl Acad Sci USA 1992; 89:7645-49; PMID:1502176; http://dx.doi.org/ 10.1073/pnas.89.16.7645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jonuscheit M, Martusewitsch E, Stedman KM, Schleper CA. Reporter gene system for the hyperthermophilic archaeon Sulfolobus solfataricus based on a selectable and integrative shuttle vector. Mol Microbiol 2003; 48:1241-52; PMID:12787352; http://dx.doi.org/ 10.1046/j.1365-2958.2003.03509.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


