Table 1.
Genotypes for the nuclear family are reported for polymorphic SNPs (CanFam2) representing each of the 10 candidate genes
| Gene (Chr) | SNP position | 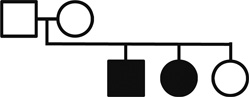 |
||||
|---|---|---|---|---|---|---|
| TNNT1 (1) | 105264488 | AG | AG | GG | GG | AG |
| 105432142 | GG | AG | AG | AG | GG | |
| 105598975 | GG | CG | CG | CG | GG | |
| 106497529 | AG | AG | AA | AA | AG | |
| ACTA1 (4) | 12713391 | AG | AA | AG | AG | AA |
| 12875911 | AG | AA | AG | AG | AA | |
| TPM3 (7) | 45997866 | CC | AC | CC | CC | CC |
| 46022207* | AT | AA | AA | AT | AT | |
| 46034262* | AC | AA | AA | AC | AC | |
| 46049585* | AG | AG | AA | AG | AG | |
| 46303991 | CC | AC | CC | CC | CC | |
| CFL2 (8) | 16507437 | CC | AC | CC | AC | CC |
| 16587360 | GG | AG | GG | AG | GG | |
| TPM2 (11) | 55241525* | AC | CC | CC | AC | AC |
| 55248320* | AA | AG | AA | AG | AA | |
| NEB (19) | 55761948* | GG | AG | GG | GG | GG |
| 55777049* | GG | AG | GG | GG | GG | |
| 55824838* | AG | AG | AA | AA | AG | |
| LMOD3 (20) | 25536336 | AG | AA | AA | AA | AG |
| 25605665 | AA | GG | AG | AG | AG | |
| 25625642 | AG | AA | AA | AA | AG | |
| KLHL40 (23) | 14709678 | AC | AA | AC | AC | AC |
| 14867876 | AG | GG | AG | AG | AG | |
| KBTBD13 (30) | 32370798 | AG | GG | GG | GG | AG |
| 32443189 | AG | GG | GG | GG | AG | |
| 32458624 | AC | AA | AA | AA | AC | |
| KLHL41 (36) | 17244342 | AG | AA | AA | AA | AG |
| 17280474 | AG | GG | AG | AG | GG | |
When possible, SNPs lying within the gene are used (asterisks); otherwise, the most proximal flanking SNPs are reported. When present, flanking SNPs (within 1 Mb) for which the affected dogs are homozygous are also shown. SNPs fitting a recessive pattern of inheritance are highlighted in bold
