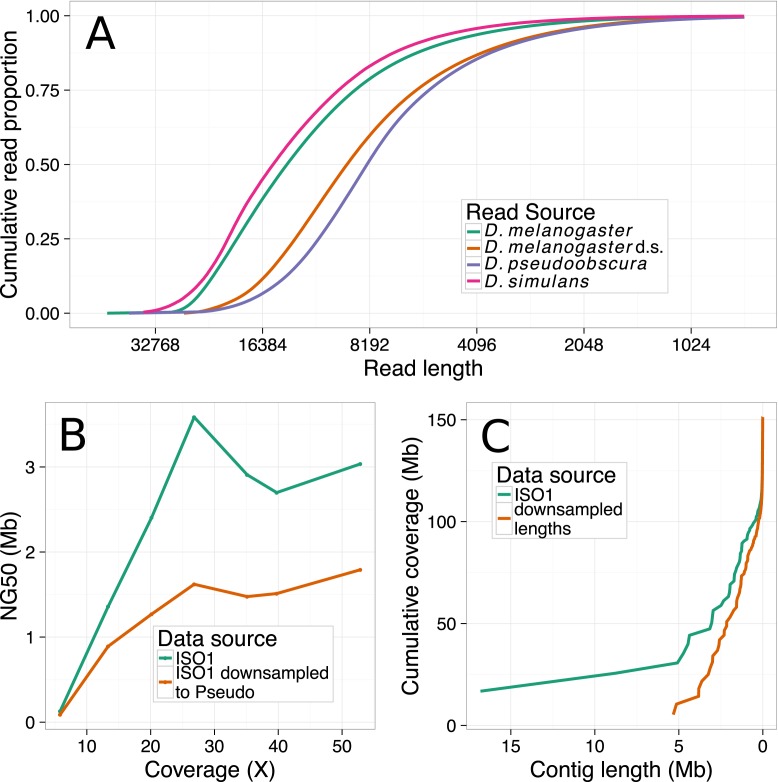
Figure 2.
(A) The cumulative read length of various data sets, where Drosophila melanogaster refers to the original ISO1 dataset, Drosophila pseudoobscura refers to a publicly available D. pseudoobscura dataset with a shorter average read length, D. melanogaster d.s. refers to the D. melanogaster data, downsampled to have read lengths resembling the D. pseudoobscura dataset and Drosophila simulans is a D. simulans dataset sequenced using our DNA preparation technique. (B) A plot of NG50 versus coverage of hybrid assemblies, as in Figure 5. This plot depicts the effect of reduced read length on NG50, while holding read quality and coverage constant. (C) Cumulative contig length distribution of 53× of PacBio only assemblies created with the original ISO1 reads and the ISO1 reads downsampled to resemble Pseudoobscura. Contig lengths in the shorter/downsampled reads assembly are considerably shorter than the contigs in the original reads assembly.