Figure 8.
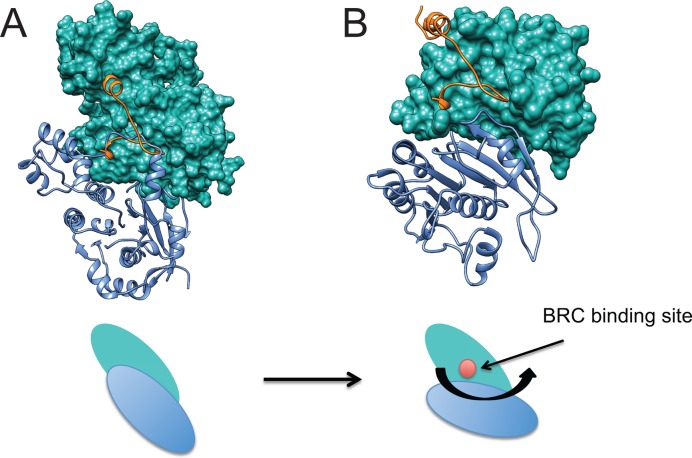
Re-orientation of RAD51 subunits during transition to an inactive filament may expose a filament-inhibitory binding site for the Phe-X-X-Ala module in the BRC repeats. Comparative modelling of the active presynaptic RAD51 filament using inactive ADP-bound RecA as a template was carried out as described in ‘Materials and Methods’ section to create a hypothetical model for an inactive RAD51 filament. Close similarity has been shown between the crystal structures of active versus ADP-bound RecA protomers (50) (RMSD < 0.8Å), and between the core catalytic domains of RecA and RAD51 (11) (RMSD < 1Å). Our modelling of the inactive RAD51 filament using RecA as a template therefore makes the minimal assumption that the structural changes underlying the active-inactive transformation are also similar. (A) Two protomers from our EM structure for the active presynaptic RAD51 filament are displayed, one in blue ribbon representation and another in green surface representation. A region of ∼99 residues from the N-terminus of both protomers was omitted, as described in the text, because this region is absent from RecA. BRC4 is shown as an orange ribbon. The cartoon below the structural model represents the relative orientation of the two RAD51 protomers in the active filament, which obscures the binding site for BRC4. (B) Two protomers from our comparative model for the inactive RAD51 filament are displayed with BRC4 using the same colour code as in the previous panel. Rotation of the protomers by ∼15° from their counterparts in the active filament exposes the BRC4 binding site (cartoon). This new conformation is free of apparent atomic clashes, speaking to its plausibility.