Figure 3.
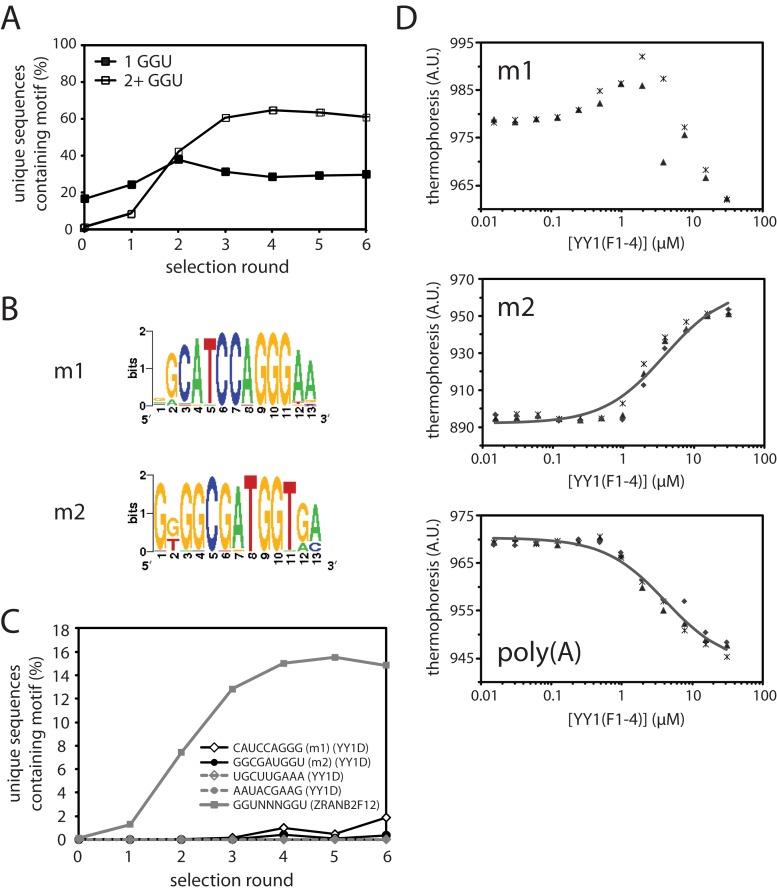
SELEX analysis of YY1 and ZRANB2(F12). (A) Enrichment of sequences containing one (solid squares) or two or more (open squares) GGU sites in the ZRANB2(F12) SELEX. (B) Weblogos of the two most highly enriched motifs identified by MERMADE from round 6 of the YY1D SELEX dataset. (C) Enrichment over successive rounds of selection of motifs m1 and m2 in YY1D SELEX libraries, and also of a representative double-GGU motif in ZRANB2(F12) libraries, as a percentage of the number of unique sequences recovered in each round. For comparison, the percentage of two other 9-mers in each round of YY1D are also shown (dashed lines). (D) Binding of YY1(F1–4) to fluorescent m2/polyA RNA in MST assays. Data points from independent titrations are shown, fitted to a 1:1 binding model.