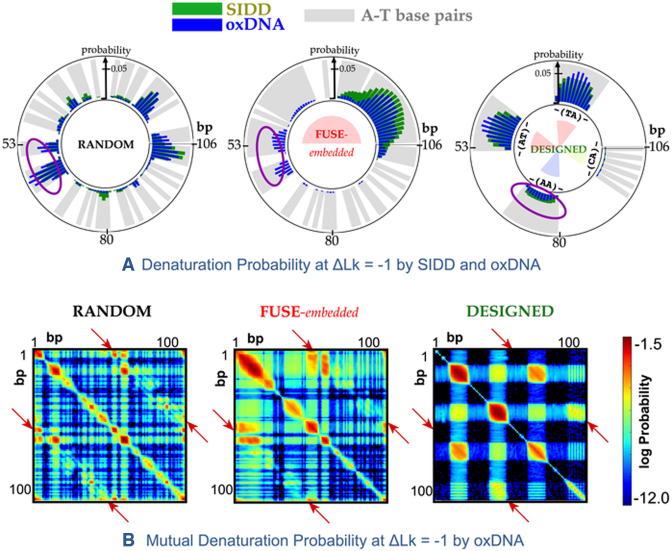
Figure 2.
(A) Sequence-dependent denaturation probability observed in negatively supercoiled DNA minicircles at ΔLk ≈ −1 of the RANDOM, FUSE-embedded, and DESIGNED sequences. This probability was calculated using the SIDD model (green) and oxDNA simulation (blue). A-T base pairs are shaded in grey. Secondary denaturation sites due to the cooperative long-ranged kinking predicted by oxDNA simulations are circled. (B) 2D colour plots of the log of the probability for a given pair of base pairs to be simultaneously denatured in ΔLk ≈ −1 minicircles, calculated with oxDNA for the same three sequences. Unsampled states are coloured black. Denaturation is enhanced at specific sites separated by ∼180° (e.g. situated antipodally) along the minicircle, as demonstrated by the off-diagonal peaks in the mutual denaturation probability profile (indicated with red arrows).