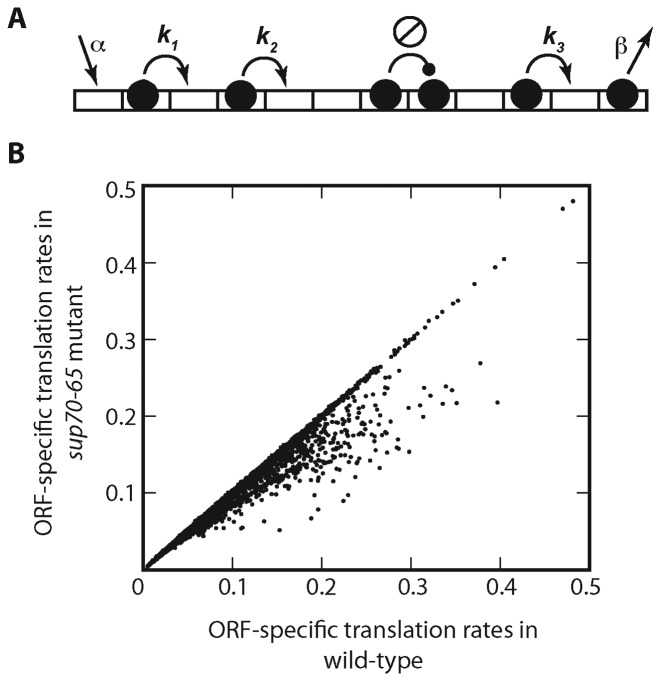
Figure 2.
Modelling ribosome flux on the total yeast transcriptome identifies the targets of glutamine 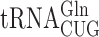 regulation. (A) Mathematical modelling of ribosome flux along each mRNA employed a Totally Asymmetric Simple Exclusion Process (TASEP) in which ribosomal particles of 9-codon width join a lattice representing the mRNA with rate α, and translate the lattice, codon by codon using 2-state dynamics. These are defined by cognate tRNA finding rate ki, dictated by tRNA abundance, and translocation rate γ for ribosomes charged with the cognate tRNA, not shown in the figure (67). Ribosome then terminate with rate β. (B) The TASEP model was used to simulate in turn each of the 5500 yeast mRNAs using published initiation and stepping rates (29). Simulations were performed using wild-type tRNA concentrations, and again using a concentration of
regulation. (A) Mathematical modelling of ribosome flux along each mRNA employed a Totally Asymmetric Simple Exclusion Process (TASEP) in which ribosomal particles of 9-codon width join a lattice representing the mRNA with rate α, and translate the lattice, codon by codon using 2-state dynamics. These are defined by cognate tRNA finding rate ki, dictated by tRNA abundance, and translocation rate γ for ribosomes charged with the cognate tRNA, not shown in the figure (67). Ribosome then terminate with rate β. (B) The TASEP model was used to simulate in turn each of the 5500 yeast mRNAs using published initiation and stepping rates (29). Simulations were performed using wild-type tRNA concentrations, and again using a concentration of 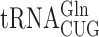 reduced to 25% of normal levels to mimic the sup70-65 mutant as determined experimentally (6). Translation rates for each mRNA under the two conditions are plotted.
reduced to 25% of normal levels to mimic the sup70-65 mutant as determined experimentally (6). Translation rates for each mRNA under the two conditions are plotted.