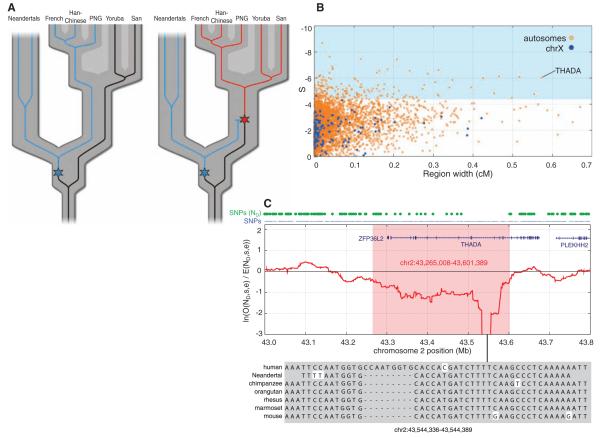
Fig. 4.
Selective sweep screen. (A) Schematic illustration of the rationale for the selective sweep screen. For many regions of the genome, the variation within current humans is old enough to include Neandertals (left). Thus, for SNPs in present-day humans, Neandertals often carry the derived allele (blue). However, in genomic regions where an advantageous mutation arises (right, red star) and sweeps to high frequency or fixation in present-day humans, Neandertals will be devoid of derived alleles. (B) Candidate regions of selective sweeps. All 4235 regions of at least 25 kb where S (see SOM Text 13) falls below two standard deviations of the mean are plotted by their S and genetic width. Regions on the autosomes are shown in orange and those on the X chromosome in blue. The top 5% by S are shadowed in light blue. (C) The top candidate region from the selective sweep screen contains two genes, ZFP36L2 and THADA. The red line shows the log-ratio of the number of observed Neandertal-derived alleles versus the number of expected Neandertal-derived alleles, within a 100 kilobase window. The blue dots above the panel indicate all SNP positions, and the green dots indicate SNPs where the Neandertal carries the derived allele.