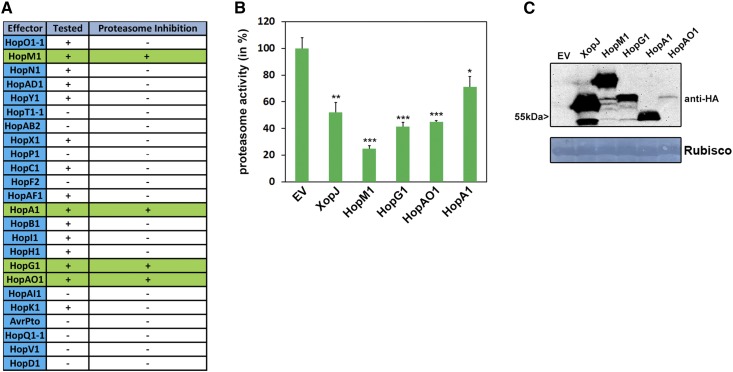
Figure 6.
T3Es from Pst suppress proteasome activity in N. benthamiana. A, Table of candidate effectors tested for their ability to modulate proteasome function. T3Es tested in proteasome activity measurements are indicated by +, and those not tested are indicated by −. T3Es suppressing proteasome activity are highlighted in green. B, Transient expression of T3Es HopM1, HopG1, HopAO1, and HopA1 in N. benthamiana suppresses proteasome activity. Proteasome activity is shown in N. benthamiana leaves following transient expression of T3Es XopJ, HopM1, HopG1, HopA1, HopAO1, or empty vector control (EV). Relative proteasome activity in total protein extracts was determined by monitoring the breakdown of the fluorogenic peptide Suc-LLVY-AMC at 30°C in a fluorescence spectrophotometer. The empty vector control was set to 100%. Data represent means ± sd (n = 3). Asterisks indicate statistical significance (*, P < 0.05; **, P < 0.01; and ***, P < 0.001) determined by Student’s t test (compared with the empty vector control). C, Protein extracts from N. benthamiana leaves transiently expressing T3Es tagged with HA and empty vector at 48 h post inoculation. Equal volumes representing approximately equal protein amounts of each extract were immunoblotted, and proteins were detected using anti-HA antiserum. Amido Black staining served as a loading control. Proteasome activity measurements were carried more than three times with these T3Es.