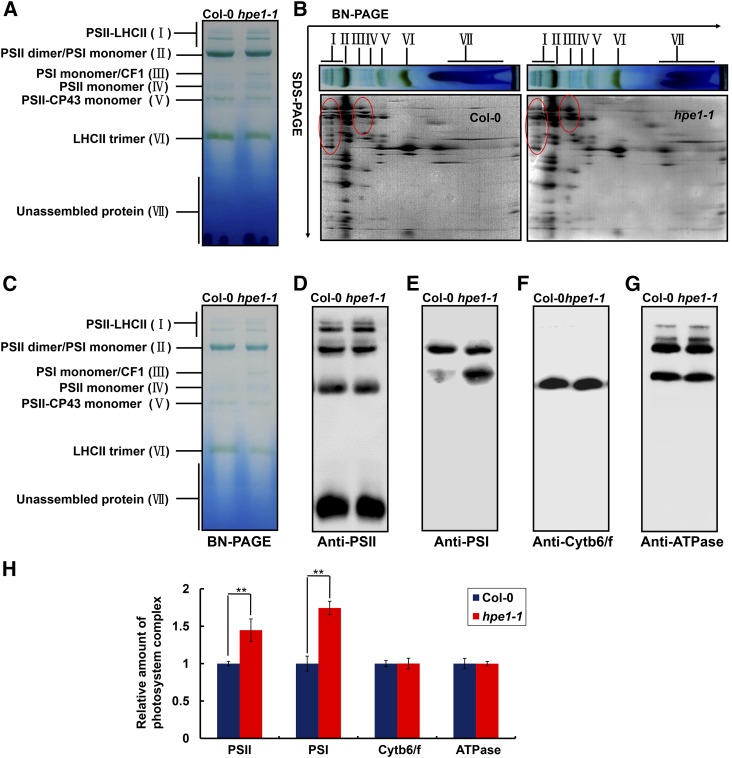
Figure 7.
Analysis of photosystem complexes from the wild-type and hpe1 mutant plants. A, BN-PAGE analysis of chlorophyll-protein complexes. Equal thylakoid membranes (10 μg of chlorophyll) from the leaves of the wild type and hpe1-1 mutants were solubilized by treatment with 1% (w/v) dodecyl β-d-maltoside and separated by BN-PAGE. The assignments of the macromolecular protein complexes of thylakoid membranes indicated at left were identified according to Jin et al. (2014). B, Two-dimensional (2D) BN/SDS-PAGE fractionation of thylakoid membrane protein complexes. After separation in the first dimension on a nondenaturing gel, the protein lanes were applied to a denaturing 2D gel followed by Coomassie Blue staining. The proteins with different abundance between hpe1 mutants and wild-type plants are circled in red. The PSII and PSI proteins, which appeared more in hpe1 mutants, are circled in red. C, BN-PAGE for immunoblot analysis (3 μg of chlorophyll). D, A representative immunoblot with anti-CP43 antiserum used to probe the PSII complex. E, A representative immunoblot with anti-PsaA antiserum used to probe the PSI complex. F, A representative immunoblot with anti-cytochrome f antiserum used to probe the cytochrome b6/f (Cytb6/f) complex. G, A representative immunoblot with anti-ATPB antiserum used to probe the ATP synthase (ATPase) complex. Three independent biological replicates for all experiments were performed, and a representative one is shown. H, Proteins immunodetected from D to G were analyzed with Phoretix 1D Software (Phoretix International). Values (means ± se; n = 3 independent biological replicates) are given as ratios to protein amounts of the wild type (Col-0) and hpe1-1 mutants. **, P < 0.01, by Student’s t test.