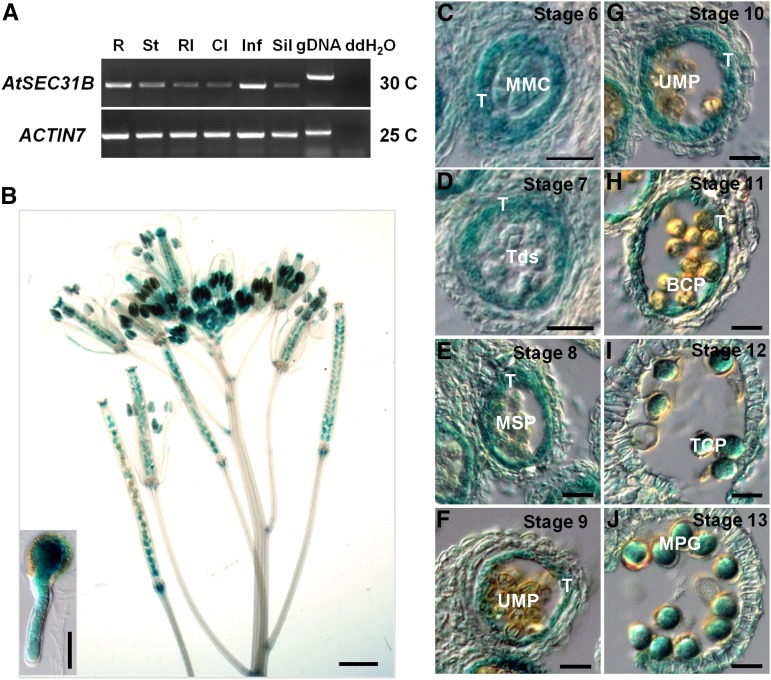
Figure 4.
Expression analysis of Arabidopsis SEC31B. A, Expression pattern of AtSEC31B in wild-type plants by RT-PCR. Total RNA was isolated from 6-week-old wild-type plants. ACTIN7 was used as an internal control. R, Root; St, stem; Rl, rosette leaf; Cl, cauline leaf; Inf, inflorescence; Sil, silique. Genomic DNA (gDNA) as a positive control and distilled, deionized water (ddH2O) as a negative control also are presented. B, GUS expression profile in the inflorescence transformed with ProAtSEC31B:GUS. The internal view in the bottom left corner shows GUS signal in a germinating pollen tube. Bar = 1 mm; for the internal view, bar = 20 μm. C to J, Paraffin sections of anthers from plants transformed with ProAtSEC31B:GUS. GUS expression was detected continuously in tapetal cells (T) before tapetum degenerated and was expressed highly in microspore mother cells (MMC), then decreased gradually in tetrads (Tds), uninuclear microspores (UMP), and bicellular pollen grains (BCP) but increased in tricellular pollen grains (TCP) and mature pollen grains (MPG). Bars = 50 μm.