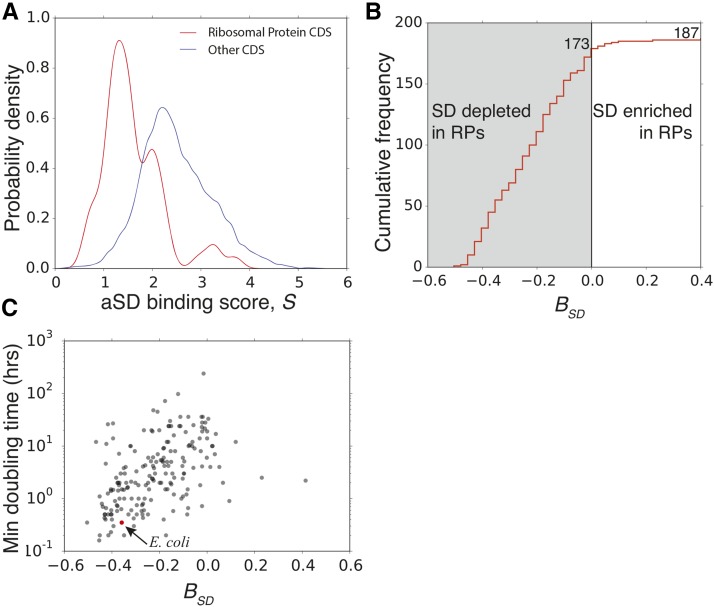
Figure 6.
Depletion of SD (Shine-Dalgarno) sequences within ribosomal protein coding genes is widespread throughout the bacterial kingdom and associated with organismal growth. (A) Distribution of aSD (anti-SD) binding scores of ribosomal protein coding sequences in E. coli, compared to that of all other protein coding sequences. We characterize SD sequence usage bias in a genome with Equation (3). (B) Distribution of genome SD bias index for 187 bacteria genomes. Ribosomal proteins have significantly lower aSD binding scores, as compared to the rest of the genome, in the majority of bacterial species. (C) SD bias is correlated with minimum generation time in 187 organisms (Spearman-rank: ). Depletion of internal-SD sequences in ribosomal protein genes is associated with faster growth. The full data table for this analysis, including organism names, growth rate, and B values, is provided as (Table S3).