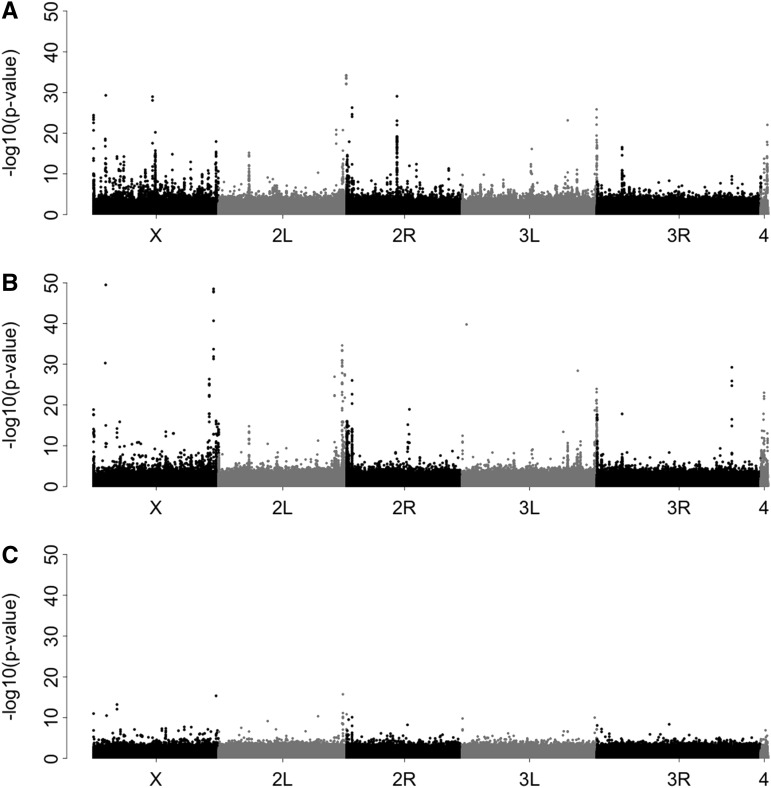
Figure 1.
Manhattan plots indicating the significance of allele frequency differences between Pool-Seq libraries when the same genomic DNA is sequenced. Two Illumina paired-end sequencing libraries with different read length and insert sizes were prepared from a pool of 250 D. simulans individuals. Reads were mapped to the reference genome, and the significance of differences in allele frequencies between the two libraries were computed (Fisher’s exact test). Although no significant allele frequency differences were expected, we found pronounced outlier peaks using bwa aln (A) or novoalign(g) (B) for mapping the reads. Importantly, outlier peaks found with these two alignment algorithms are at different genomic sites. Hence, intersecting the results of these two algorithms by plotting the lowest P-value obtained at each site removes the vast majority of outlier peaks (C).