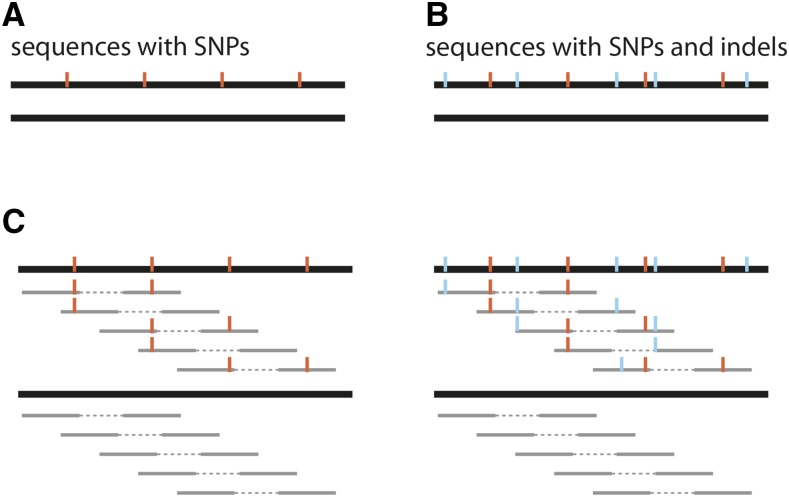
Figure 2.
Overview of simulated Pool-Seq data sets. Based on a 2 Mbp region of D. melanogaster chromosome 2R, we simulated a pair of sequences, with one sequence having a SNP (red) every 100 bp (A), and a pair of sequences with one sequence having, in addition to the SNPs, an indel (blue) with random position and length between adjacent SNPs (B). Using these sequences as templates, we simulated uniformly distributed paired ends (gray; C) resulting in SNPs with known positions and frequency (f = 0.5).