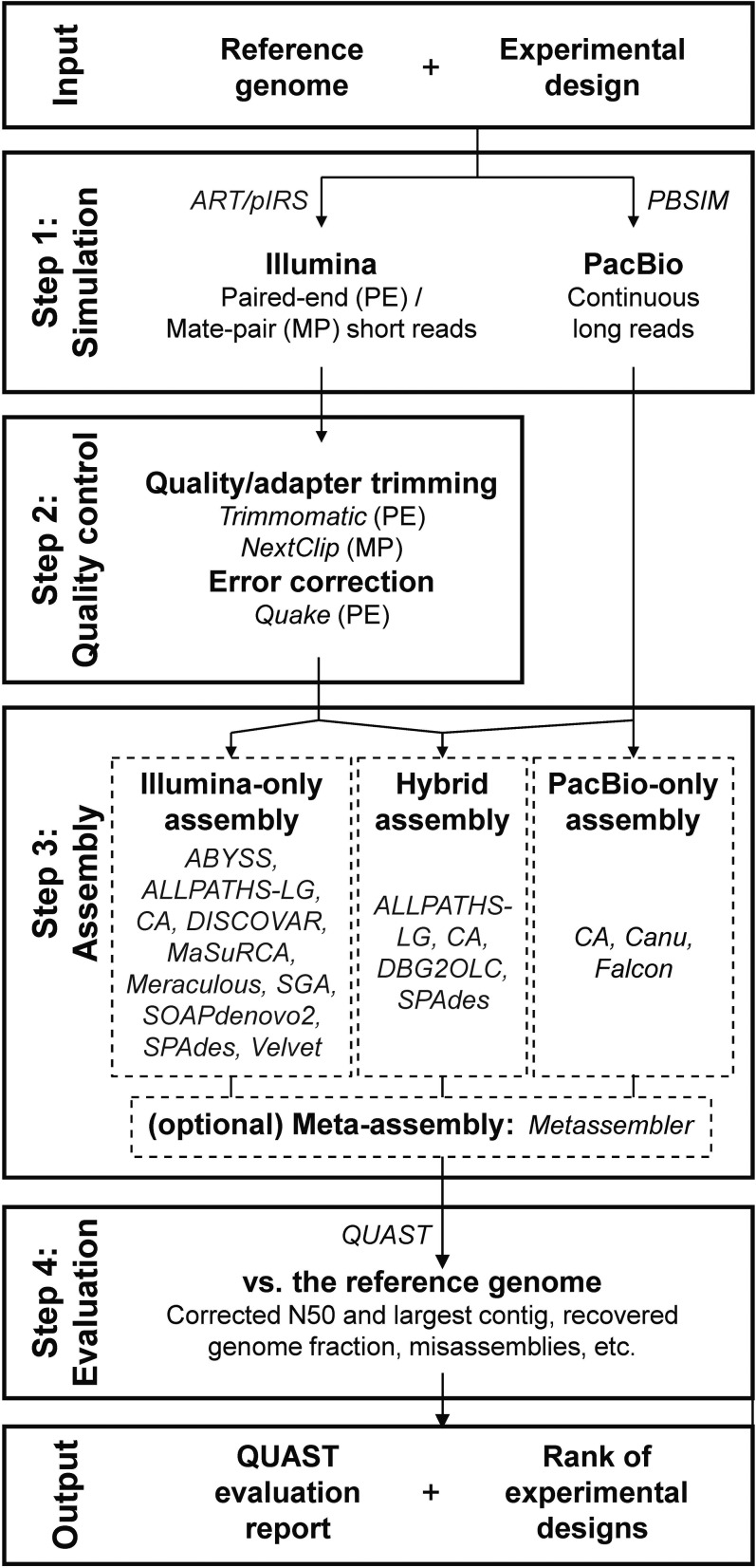
Figure 1.
iWGS workflow. A typical iWGS analysis consists of four steps: (1) data simulation (optional); (2) preprocessing (optional); (3) de novo assembly; and (4) assembly evaluation. iWGS supports both Illumina short reads and PacBio long reads, and a wide selection of assemblers to enable de novo assembly using either or both types of data. Users can start the analysis simulating data drawn from a reference genome assembly or, alternatively, use real sequencing data as input and skip the simulation step. iWGS, in silico Whole Genome Sequencer and Analyzer; MP, mate-pair; PacBio, Pacific Biosciences; PE, paired-end.