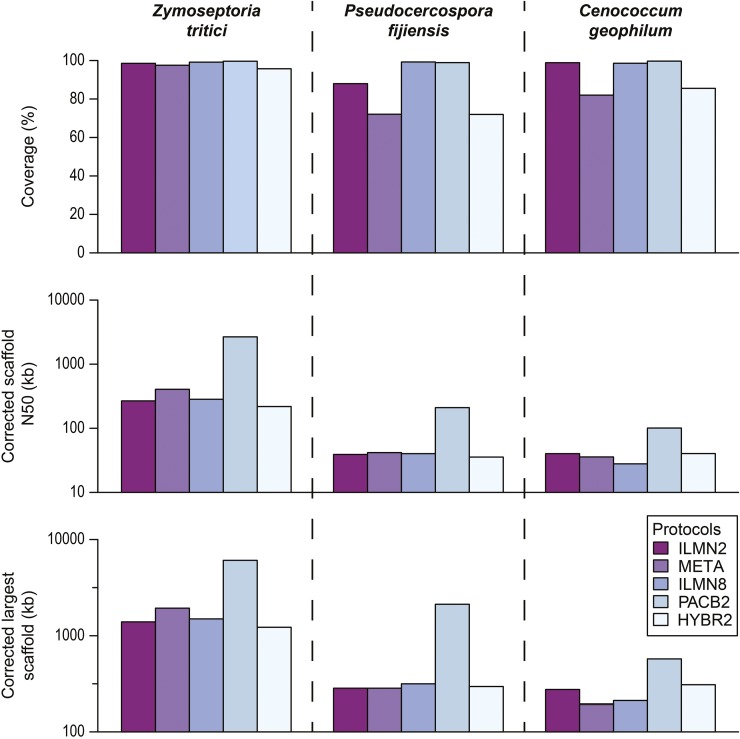
Figure 2.
Performance comparison of five representative experimental designs on three Dothideomycetes genomes. The five designs shown include three Illumina-only designs (ILMN2: ALLPATHS-LG, META: Metassembler, and ILMN8: DISCOVAR), the best performing PacBio-only design (PACB2: Canu), and the best performing hybrid design (HYBR2: DBG2OLC) for each genome. The statistics on the assembled fraction of the reference genome, scaffold N50, and largest scaffold size are all after correction for assembly errors using the reference genome as reported by QUAST in GAGE mode. By default, QUAST (in GAGE mode) corrects contigs/scaffolds by breaking them at assembly errors larger than 5 bp. Scaffold N50 and largest scaffold size are shown in log10 scale.