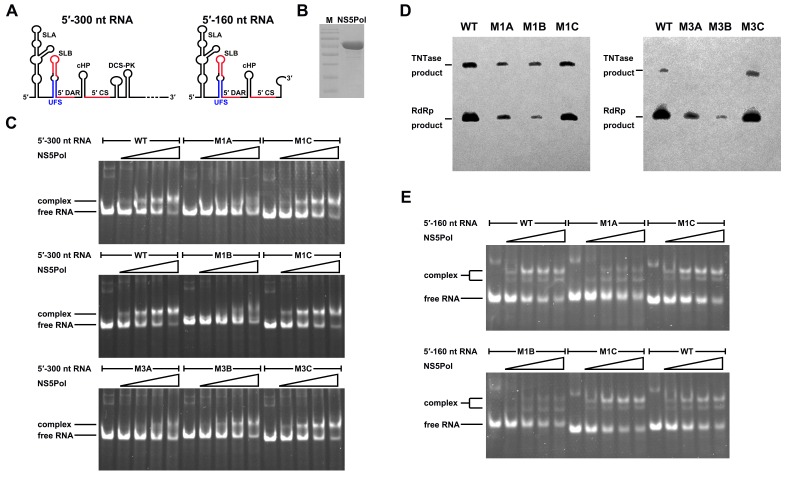
Figure 6. UFS is crucial for RdRp recruitment and de novo RNA synthesis.
(A) Simplified diagrams of DENV4 virus 5′ end RNA constructs used for RdRp binding and/or in vitro RdRp activity assays. The region corresponding to the UFS element is shown in blue, and the red regions represent the genome cyclization elements. (B) SDS-PAGE of purified recombinant NS5Pol of DENV4. (C) The binding of DENV4 NS5Pol to 5′-300 nt RNA molecules containing UFS mutations was analyzed by EMSA. No NS5Pol was present in the left first lane of each group. The NS5Pol concentrations in the reactions were approximately 2.0, 3.5, 5.0 and 7.5 μM (from left to right). (D) In vitro RdRp activity assay using 5′-160 nt RNA as a template. The reactions were incubated at 30°C for 20 min. Detection was based on the incorporation of biotin-11-CTP into the products. Left, the results of the M1 series; right, the results of the M3 series. Two bands were detectable in the blot: the upper template-sized band was generated by the terminal addition of biotin-11-CTP to the input template due to the terminal nucleotide transferase activity of NS5Pol in the presence of Mn2+, and the lower band represented the dsRNA product of de novo RNA synthesis, which was confirmed by comparison of the electrophoretic patterns of the RdRp products and annealed dsRNA molecules (Figure 6—figure supplement 2). (E) EMSA was performed for the 5′-160 nt RNA molecules containing M1 series mutations. The NS5Pol concentrations in the reactions were approximately 3.6, 7.2, 10.7 and 14.3 μM (left to right).
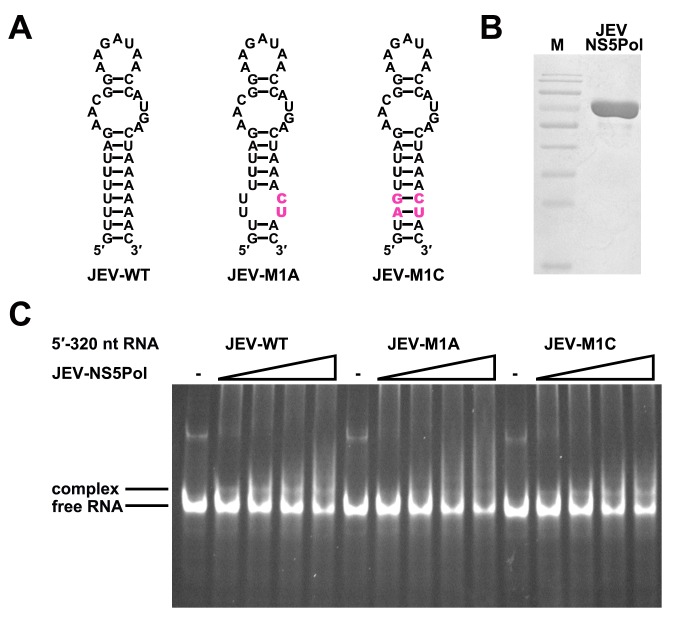
Figure 6—figure supplement 1. The UFS is required for efficient NS5 binding to 5′ RNA in JEV.
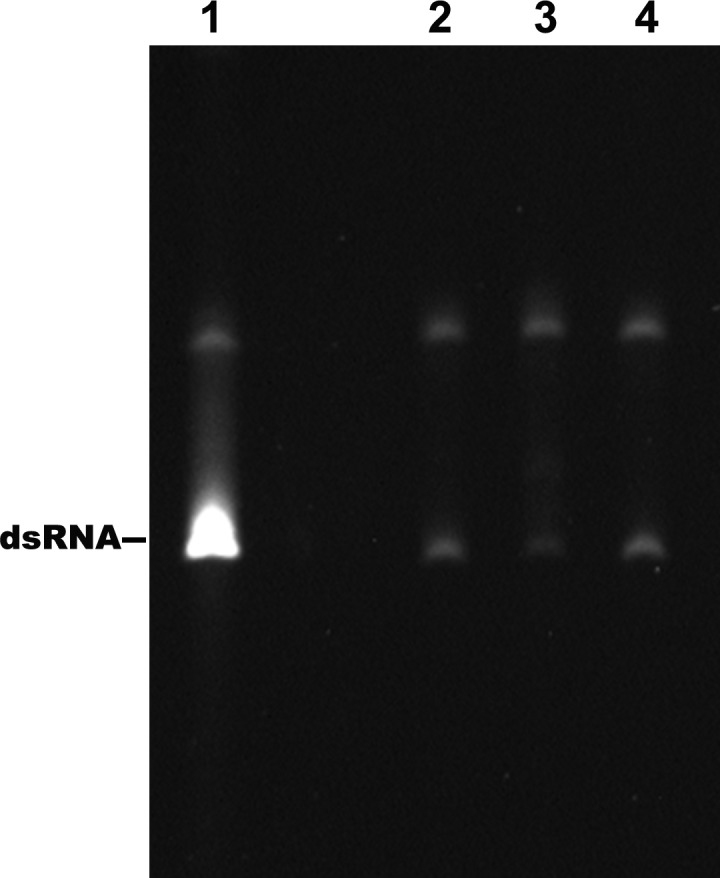
Figure 6—figure supplement 2. Identification of the products of the RdRp reactions.