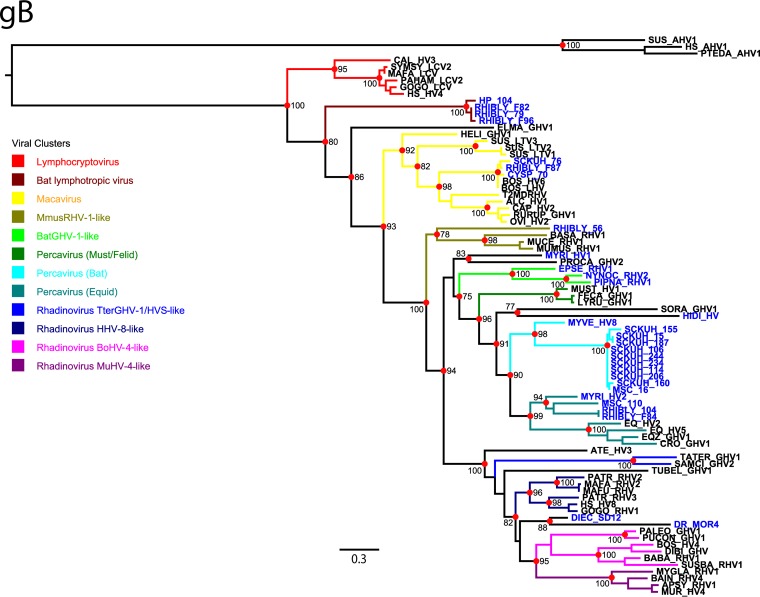
FIG 1 .
The phylogeny of gammaherpesviruses based on a 564-residue-long alignment of the glycoprotein B (gB) sequence. Maximum-likelihood tree estimated from 81 mammalian γHV sequences, including 30 viral sequences from 14 different bat species. The tree is color coded according to the major γHV clusters, while bat viral sequences are highlighted in blue. The tree was rooted with alphaherpesvirus sequences. Branch support values are shown for nodes with support values of >70% according to the Shimodaira-Hasegawa-like approximate-likelihood ratio test (SH-like–aLRT), represented by red circles. The full names for the viral isolates and their original hosts are available in Table S5 in the supplemental material. The scale bar denotes amino acid substitutions per site.