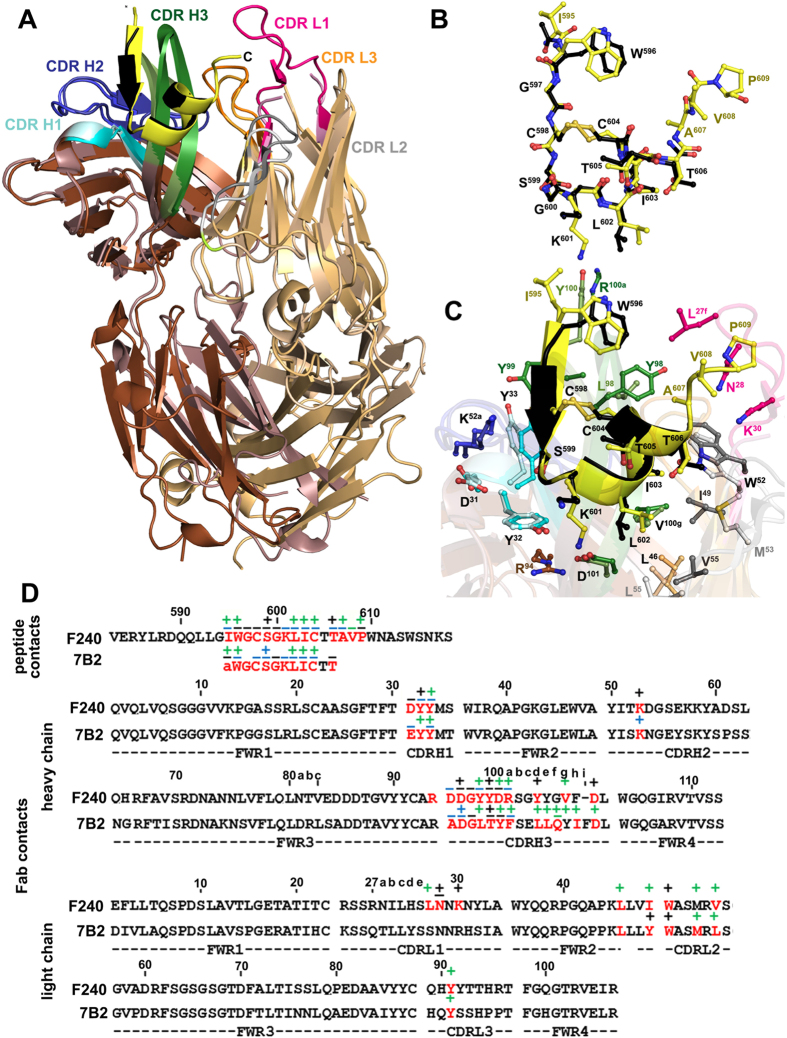
Figure 3. Structural comparison of F240- gp41583–618 and 7B2 Fab- gp41596-606 complexes.
F240 and 7B2 use the same VH3–3.11 and VLκ4.1 germline gene sequences for VH and VL domains, respectively. (A) The structure of the F240- gp41583–618 complex (dark brown/light brown) was superimposed onto the 7B2 Fab- gp41596–606 (dark pink/light pink) (PDB code 4YDV) based on gp41 peptide molecules. The CDRs are shown as dark and light shades as in Fig. 2 for F240 Fab and 7B2 Fab, respectively. The gp41 peptide is shown as a yellow and black ribbon for F240 Fab-gp41583–618 and 7B2 Fab-gp41596–606 complexes, respectively. (B) Superposition of gp41583–618 and gp41596–606 peptides bound to F240 and 7B2 Fab, respectively. Peptides were superimposed based on main chain atoms. (C) The F240 Fab-gp41583–618 and 7B2 Fab-gp41596–606 binding interfaces. Residues involved in the interactions between F240/7B2 Fab and gp41583–618/gp41596–606 peptide as defined by PISA are shown as sticks (D) Residues in F240 and 7B2 involved in the gp41 peptide binding interface. Buried residues are highlighted red. Main chain (−) and side chain (+) as defined by a 5 Å distance criteria cutoff and colored based on contact type, hydrophobic (green), hydrophilic (blue), or both (black). Framework and CDRs are indicated below the alignment. Only the variable regions of the heavy and light chains for both antibodies are shown.