Abstract
The present study aimed to investigate the differential expression and clinical significance of histone methyltransferase G9a, histone H3K9me2 and histone H3K9me1 in human brain glioma and adjacent tissue samples. It also aimed to observe the effect and mechanism of BIX-01294, as an inhibitor of methyltransferase G9a, on the proliferation, apoptosis, methylation of H3K9 and H3K27, and the acetylation in U251 glioma cells in vitro. The differential expression of methyltransferase G9a, histone H3K9me2 and histone H3K9me1 in in human brain glioma and adjacent tissues were analyzed by immunohistochemistry, a growth curve of U251 cells following treatment with BIX-01294 was determined using the MTT assay. In addition, the apoptosis percentage of U251 cells was analyzed by TUNEL assay and the expression levels of apoptosis-associated proteins, including B-cell lymphoma 2 (Bcl-2), Bcl-2-associated X protein (Bax), caspase-9 and caspase-3, and the acetylation of histones, including H3K27me1, H3K27me2 and H3 in U251 were analyzed by western blot following BIX-01294 treatment. The positive rate of G9a in glioma tissues was 86% (43/50), which was significantly different from 42% (21/50) in adjacent tissues (P<0.01). The positive rate of H3K9me2 in glioma tissues was 82% (41/50), which was significantly different from 38% (19/50) in adjacent tissues (χ2=18.38; P<0.01). The expression of G9a and H3K9me2 were associated with the World Health Organization (WHO) glioma grade. The positive rate of H3K9me1 in glioma tissues was 54% (27/50) and 44% (22/50) in adjacent tissues, though this result was not significantly different (χ2=1.21, P>0.05). BIX-01294 inhibited the proliferation of U251, downregulated expression of Bcl-2, and upregulated expression of Bax, caspase-3 and caspase-9, and induced apoptosis of U251. BIX-01294 downregulated H3K9me1, H3K9me2, H3K27me1 and H3K27me2, however, it did not affect the acetylation of H3K9me3 and H3. High expression of G9a and H3K9me2 in glioma tissue samples was associated with the WHO grade, which indicated that G9a and H3K9me2 may promote generation and development of glioma. BIX-01294 inhibited proliferation and induced apoptosis of glioma cells, changes in methylation of H3K9 and H3K27 resulting in conformational changes of chromosome may be an underlying mechanism. BIX-01294 may be a potential novel therapeutic agent in the treatment of glioma.
Keywords: G9a, glioma, histone lysine demethylase, histone methylation, apoptosis, epigenetics
Introduction
Histone methylation is conducted by the coordination of specific histone methyltransferase and demethylase, as a reversible dynamic process. The abnormal expression of histone methyltransferase and demethylase are important in the generation and development of glioma. It has been demonstrated that there were marked changes to expression of histone methyltransferases, including histone-lysine-N-methyltransferase SETD7, histone-lysine N-methyltransferase 2A and myeloid/lymphoid or mixed-lineage leukemia 4, and to demethylases, including lysine-specific demethylase 2A and lysine-specific demethylase 2B in glioma cells (1). G9a, as the important methyltransferase of euchromatin, is important in the regulation of epigenetic inheritance. Huang et al (2) demonstrated that G9a and G9a-like protein 1 (GLP) may methylate tumor suppressor gene TP53, resulting in methylated TP53, which has no activity (2). H3K9 methylation induced by G9a may hold the ability of transcriptional inhibition. However, H3K9 methylation is important in transcriptional silencing, X chromatin deactivation and tumor generation and development. BIX-01294 is an artificial inhibitor of G9a with high selectivity. Kondo et al (3) demonstrated that following BIX-01294 treatment, the activity of G9a and H3K9 methylation decreased, leading to a decrease in chromosome stability, an increase in the apoptosis of tumor cells and cycle arrest (3).
The present study investigated the differential expression and clinical significance of histone methylase G9a, histone H3K9me2 and histone H3K9me1 in human brain glioma and adjacent tissues. By observing the influence of BIX-01294 on biological changes, histone methylation and acetylation in U251 cells, a possible mechanism was also investigated.
Materials and methods
Subjects
All the patients (20 males and 21 females; mean age, 34.6) were hospitalized for neurosurgery in Zhangzhou Affiliated Hospital of Fujian Medical University (Zhangzhou, China) between January 2010 to June 2013. All the patients were diagnosed with glioma and underwent complete follow-up treatment, without any therapy received prior to surgery. The data, including pathological specimens and clinical materials were collected. According to the World Health Organization (WHO) classification standard of central nervous system neoplasms (4), the patients were graded into WHO I (8 patients), WHO II (21 patients), WHO III (15 patients) and WHO IV (6 patients). The tumor specimens and samples from the junctional area between the tumor and normal brain tissue were collected to serve as the experimental group and control group. Written informed consent was obtained from the participants prior to the start of the study. This study was approved by the Ethics Committee of Zhangzhou Affiliated Hospital of Fujian Medical University.
Agents
BIX-01294 was obtained from Sigma-Aldrich (Merck Millipore, Darmstadt, Germany). Fetal bovine serum (FBS) was obtained from Hangzhou Sijiqing Bioengineering Material Co., Ltd., Hangzhou, China. Antibodies against G9a (cat. no. 09-071) H3K9me1 (cat. no. 07-450), H3K9me2 (cat. no. 16–187), H3K9me3 (cat. no. 07–442), H3K27me1 (cat. no. 07–448), H3K27me2 (cat. no. 07–452), Acteylated (Act)-H3 (cat. no. 07–677-I), caspase-9 (cat. no. 05–672), caspase-3 (cat. no. 05–654), B-cell lymphoma 2 (Bcl-2; cat. no. 05–826), Bcl-2-associated X protein (Bax; cat. no. AB2915) and β-actin (cat. no. 04–1116) were obtained from Upstate Biotechnology, Inc. (Lake Placid, NY, USA) and used at dilutions between 1:200-1:500. Goat anti-rabbit (cat. no. sc-3837) and goat anti-mouse (cat. no. sc-395758) antibodies were obtained from Santa Cruz Biotechnology, Inc. and used at dilutions 1:2,000–1:5,000.
Cell culture
The U251 human glioma cell line was obtained from Shanghai Institute for Biological Sciences, Chinese Academy of Sciences (Shanghai, China). RPMI 1640 was obtained from Gibco (Thermo Fisher Scientific Inc., Waltham, MA, USA), containing 10% FBS and 2 mM L-glutamine. U251 cells were cultured at 37°C with saturated humidity and 5% CO2. The cells were passaged every 3 to 4 days, with 0.25% trypsin digesting for 2–3 min followed by seeding of the cell suspension to the required concentration. Prior to seeding, the activity of U251 cells was detected by trypan blue staining.
Detection of G9a, H3K9me2 and H3K9me1 expression in glioma and adjacent tissues
The proteins were detected using the streptavidin-peroxidase method (5). G9a (1:200), H3K9me2 (1:300) and H3K9me1 (1:300) antibodies were used. The results of the immunohistochemistry were judged by the semi-quantitative integral method, according to the sum of the color strength and the percentage of stained cells. Color strength grade: Without staining, 0; weak staining, 1; moderate staining, 2; and strong staining, 3. Coloring percentage: Without staining, 0; <25%, 1; 25–50%, 2; 50–75%, 3; and >75%, 4. Sum of the two: 0, negative; 1–2, + (negative); 3–4, ++ (positive); 5–6, +++ (positive); and 7, ++++ (positive). All the data were evaluated by experienced pathologists using double blinding.
MTS assay to detect the cell growth curve following different concentrations of BIX-01294
Cells in the logarithmic growth phase were seeded in a 96-well plate (Costar; Corning Incorporated, Corning, NY, USA) at a concentration of 1.0×105/ml (100 µl each well). BIX-10294 was added and the concentration adjusted to 0, 1, 2, 4 and 8 µmol/l with 6 wells for each group. After 24 h, 48 h and 72 h at 37°C, 100 µl MTS (5 mg/ml; Sigma-Aldrich; Merck Millipore) was added and incubated for 4 h at 37°C. The cells were centrifuged at 800 × g for 5 min and the supernatant discarded. DMSO (100 µl; Sigma-Aldrich; Merck Millipore) was added and mixed fully. The absorbance (value A) was detected at wavelengths of 492 and 630 nm using a microplate reader. The cell proliferation rate was calculated according to the 0 µM (blank). Cell proliferation rate (%)= (Aexperiment - Ablank) / (Acontrol-Ablank) × 100%. Repeated totally 3 times.
Apoptosis detection by TUNEL method
U251 cells in the logarithmic growth phase were seeded on 6-well plates at 1×106 cells per well, a cover glass was also placed in each well. After 24 h, BIX-01294 was added, the concentration was adjusted to 0, 2, 4 and 8 µmol/l and the cells were incubated for 24 h at 37°C. The TUNEL assay was conducted according to the manufacturer's protocols (DeadEnd™ Fluorometric TUNEL System; Promega Corporation, Madison, WI, USA) and cells were imaged using light microscopy.
Western blotting to detect the changes of apoptosis-associated proteins following BIX-01294 treatment
The cells were centrifuged and collected following treatment for 24 h. The cells were washed twice with PBS. Lysis buffer (100 µl) and 1 µl enzyme inhibitor were added to 1×106 cells on ice for 30 min. The cells were then centrifuged at 10,000 × g, at 4°C for 10 min to extract the protein and protein quantification was conducted using the bicinchoninic acid method. The proteins (20 µg per lane) were separated by 12% SDS-PAGE gel electrophoresis and transferred to a polyvinylidene fluoride membrane (EMD Millipore, Billerica, MA, USA). Following transfer, membranes were blocked with 5% non-fat milk for 1 h at room temperature, and incubated with primary antibodies, H3K9me (1:500), H3K9me2 (1:500), H3K9me3 (1:500), H3K27me (1:1,000), H3K27me2 (1:1,000), Act-H3 (1:2,000), caspase 9 (1:600), caspase 3 (1:600), Bcl-2 (1:500) and Bax (1:500), at 4°C overnight. The membrane was then washed with Tris-buffered saline and goat anti-mouse secondary antibodies (1:5,000) were added and incubated for 1 h at room temperature. Following washing with TBS, the proteins were analyzed by chemiluminescence using β-actin as a loading control. The images were analyzed by AlphaDigiDoc imaging analysis software (version 7.1; Alpha Innotec, Kasendorf, Germany).
Statistical analysis
Data are presented as the mean ± standard deviation. All the data were analyzed by SPSS 13.0 software (SPSS, Inc., Chicago, IL, USA). Homogeneity of variance and a normality test were conducted and one-way analysis of variance was used to analyze the results. P<0.05 was considered to indicate a statistically significant difference.
Results
G9a is differentially expressed in glioma and adjacent tissues
In glioma tissues, the positive rate of G9a was 86% (43/50), which was significantly higher than that in the adjacent tissues, 42% (21/50; χ2=19.14, P<0.01; Table I). In grade I glioma tissues, G9a expression included 4 classified as ++, 1 classified as +++ and 0 classified as ++++, grade II including 8 classified as ++, 7 classified as +++ and 3 classified as ++++, grade III including 5 classified as ++, 6 classified as +++ and 3 classified as ++++, and grade IV including 2 classified as +++ and 4 classified as ++++. There were significant differences in the differential expression among different grades of glioma (F=37.52, P<0.01). The higher WHO grade was associated with higher G9a expression intensity (Table II).
Table I.
Differential expression of histone methylase G9a in glioma and adjacent tissues.
| Group | Cases (n) | Positive (n) | Positive rate (%) | P |
|---|---|---|---|---|
| Glioma tissue | 50 | 43 | 86.00 | |
| Adjacent tissue | 50 | 21 | 42.00 | <0.01 |
Table II.
Differential expression of histone methylase G9a in different glioma tissue.
| WHO | |||||
|---|---|---|---|---|---|
| Positive stage | Cases (n=) | Positive (n=) | ++ | +++ | ++++ |
| I | 8 | 5 | 4 | 1 | 0 |
| II | 21 | 18 | 8 | 7 | 3 |
| III | 15 | 14 | 5 | 6 | 3 |
| IV | 6 | 6 | 0 | 2 | 4 |
WHO, World Health Organization.
H3K9me2 is differentially expressed in glioma and adjacent tissues
The positive rate of H3K9me2 in glioma tissues was 82% (41/50), which was significantly increased from 38% (19/50) in adjacent tissues (χ2=18.38, P<0.01; Table III). In grade I glioma tissue, H3K9me2 expression included 3 classified as ++, 1 classified as +++ and 0 classified as ++++, grade II including 7 classified as ++, 5 classified as +++ and 6 classified as ++++, grade III including 4 classified as ++, 5 classified as +++ and 4 classified as ++++, and grade IV including 3 classified as +++ and 3 classified as ++++. There were significant differences in the differential expression among different grades of glioma (F=30.28, P<0.01). Higher WHO grade was associated with higher H3K9me2 expression intensity (Table IV).
Table III.
Differential expression of H3K9me2 in glioma and adjacent tissues.
| Group | Cases (n) | Positive (n) | Positive rate (%) | P |
|---|---|---|---|---|
| Glioma tissue | 50 | 41 | 82.00 | |
| Adjacent tissue | 50 | 19 | 38.00 | <0.01 |
Table IV.
Differential expression of H3K9me2 in different glioma tissue samples.
| Positive | |||||
|---|---|---|---|---|---|
| WHO stage | Cases (n) | Positive (n) | ++ | +++ | ++++ |
| I | 8 | 4 | 3 | 1 | 0 |
| II | 21 | 18 | 7 | 5 | 6 |
| III | 15 | 13 | 4 | 5 | 4 |
| IV | 6 | 6 | 0 | 3 | 3 |
WHO, World Health Organization.
H3K9me1 is differentially expressed in glioma and adjacent tissues
The positive rate of H3K9me1 in in glioma tissues was 54% (27/50) which was not significantly different from the adjacent tissues were the positive rate was 44% (22/50) in adjacent tissues (χ2=1.21, P>0.05; Table V). In grade I glioma tissues, H3K9me2 expression included 1 classified as ++, 1 classified as +++ and 2 classified as ++++, grade II including 4 classified as ++, 5 classified as +++ and 3 classified as ++++, grade III including 2 classified as ++, 3 classified as +++ and 3 classified as ++++, and grade IV including 1 classified as +++ and 2 classified as ++++. There were no significant differences in the differential expression among different grades of glioma (F=1.28, P>0.05; Tables V and VI, Figs. 1–3).
Table V.
Differential expression of H3K9me1 in glioma and adjacent tissues.
| Group | Cases (n) | Positive (n) | Positive rate (%) | P |
|---|---|---|---|---|
| Glioma tissue | 50 | 27 | 54.00 | |
| Adjacent tissue | 50 | 22 | 44.00 | <0.01 |
Table VI.
Differential expression of H3K9me1 in different glioma tissue samples.
| Positive | |||||
|---|---|---|---|---|---|
| WHO | Cases (n) | Positive (n) | ++ | +++ | ++++ |
| I | 8 | 4 | 1 | 1 | 2 |
| II | 21 | 12 | 4 | 5 | 3 |
| III | 15 | 8 | 2 | 3 | 3 |
| IV | 6 | 3 | 1 | 0 | 2 |
WHO, World Health Organization.
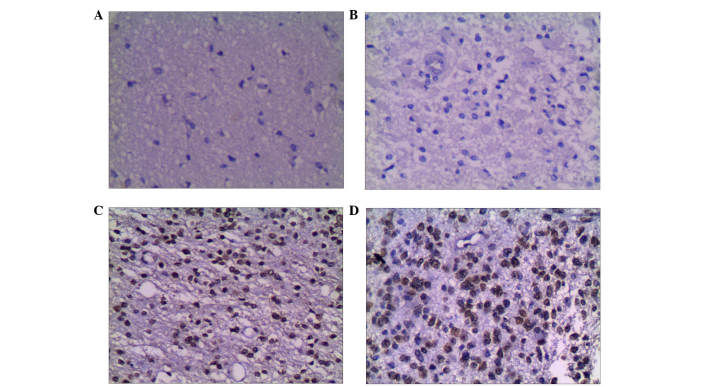
Figure 1.
Expression of G9a in glioma and adjacent tissues (Olympus BX41; magnification, ×200). (A) Negative result for G9a in adjacent tissues. (B) Negative result for G9a in grade I glioma. (C) Positive result for G9a in grade II glioma. (D) Positive result for G9a in grade IV glioma. G9a, histone methyltransferase G9a.
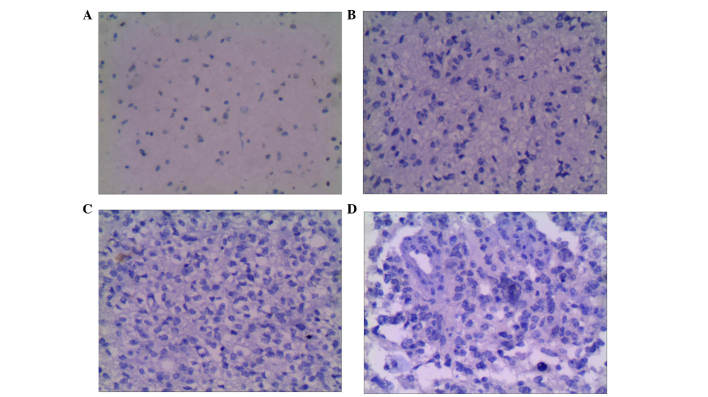
Figure 3.
Expression of H3K9me1 in glioma and adjacent tissues (Olympus BX41; magnification, ×200). (A) Negative H3K9me in adjacent tissues. (B) Negative H3K9me in grade II glioma. (C) Negative H3K9me in grade III glioma. (D) Negative H3K9me in grade IV glioma.
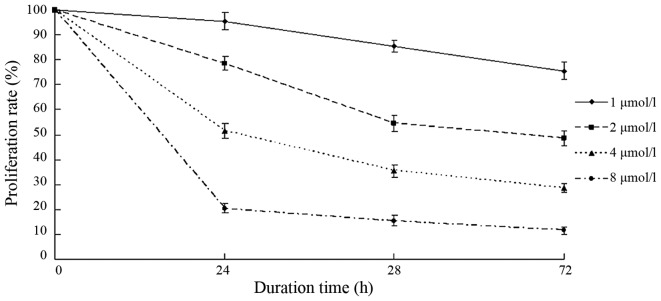
BIX-01294 inhibited the proliferation of U251 cells
Following treatment with different concentrations of BIX-01294 (1, 2, 4 and 8 µmol/l) for 24 h, the proliferation rates of U251 cells were 94.12±3.41, 78.83±2.25, 53.68±2.54 and 21.04±2.07%, respectively. As the concentration increased, the proliferation inhibition rate increased and the proliferation rate decreased (Fig. 4). After 24 h, 48 h and 72 h of BIX-01294 treatment, the proliferation of U251 cells decreased. The proliferation rate also decreased in a time-dependent manner.
Figure 4.
The changes of proliferation rate following treatment with different concentrations of BIX-01294.
BIX-01294 induced apoptosis of U251 cells
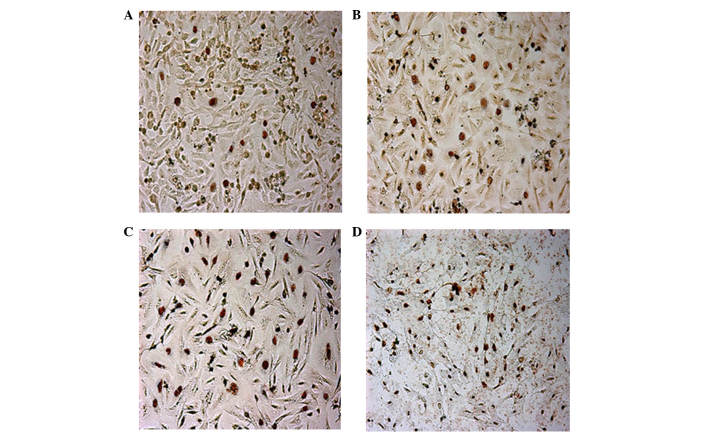
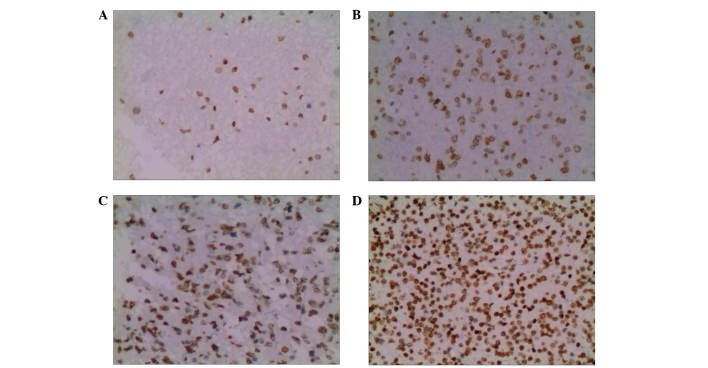
Following treatment with different concentrations of BIX-01294 (1, 2, 4 and 8 µmol/l) for 24 h, the apoptosis rates of U251 cells were 3.67±1.42, 16.42±5.18, 35.18±3.26 and 57.52±4.37%, respectively, which indicates apoptosis rates significantly increased in a concentration-dependent manner (F=32.52, P<0.01; Fig. 5).
Figure 5.
Change in of apoptosis rate following treatment with BIX-01294 for 24 h. The apoptotic cells were TUNEL positive as observed by light microscopy, and the apoptotic nuclei were stained yellow-brown and dark brown. Treatment with (A) 0 µmol/l, (B) 2 µmol/l, (C) 4 µmol/l and (D) 8 µmol/l resulted in the following changes: (A) 3.67±1.42%, (B) 16.42±5.18%, (C) 35.18±3.26% and (D) 57.52±4.37%.
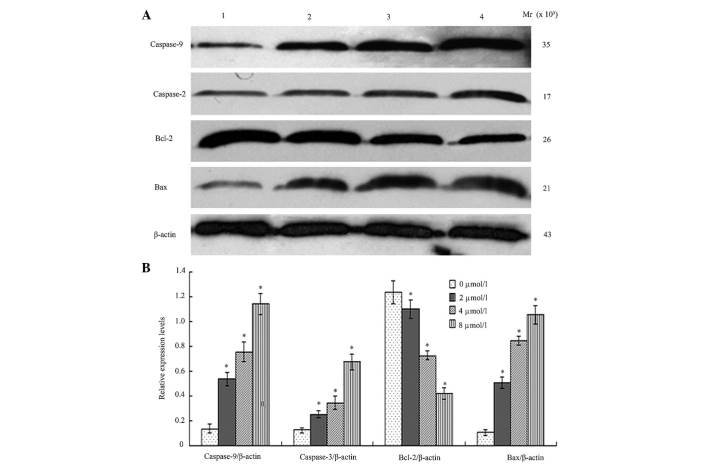
BIX-01294 altered expression levels of apoptosis-associated proteins in U251 cells
Following treatment different concentration of BIX-01294 (1, 2, 4 and 8 µmol/l) for 24 h, the proteins were analyzed by western blotting and AlphaDigiDoc image analysis. Bcl-2 expression was demonstrated to be downregulated, while Bax, caspase-9 and caspase-3 were upregulated. All the gray values of the protein bands were compared with β-actin and statistically compared to the 0 µmol/l (blank group), indicating significant differences (P<0.05, Fig. 6).
Figure 6.
Changes in expression of apoptosis-associated proteins following treatment with BIX-01294 for 24 h. (A) Lane 1, 0 µmol/l; lane 2, 2 µmol/l; lane 3, 4 µmol/l; and lane 4, 8 µmol/l. (B) Quantification of the changes to expression levels. *P<0.05 vs. 0 µmol/l.
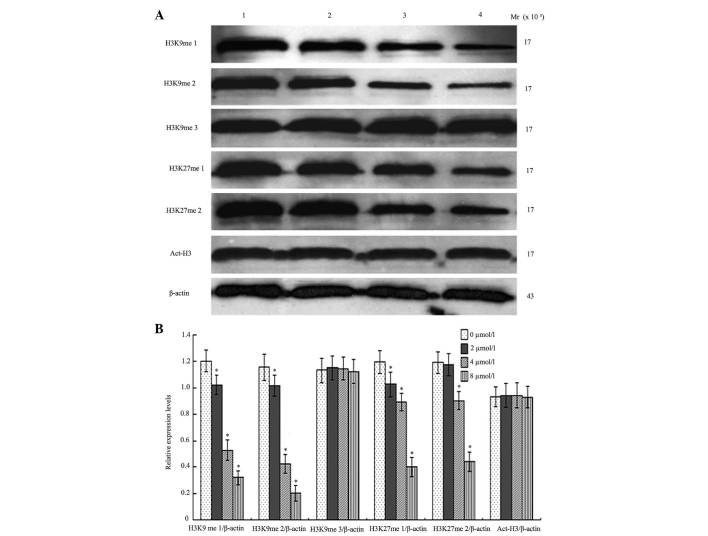
BIX-01294 affects the methylation of H3K9 and H3K27, and the acetylation of H3
Following treatment with different concentrations of BIX-01294 (1, 2, 4 and 8 µmol/l) for 24 h, the monomethylation and dimethylation of H3K9 decreased, while the trimethylation was not markedly influenced, which indicates there was activity for monomethylation and dimethylation but reduced activity for trimethylation. G9a was also demonstrated to exert an effect on the first and second methylation of H3K27. Following treatment with different concentrations of BIX-01294 (1, 2, 4 and 8 µmol/l) for 24 h, there were no marked changes to H3 acetylation (Fig. 7).
Figure 7.
Changes in histone expression following treatment with BIX-01294 for 24 h. (A) Lane 1, 0 µmol/l; lane 2, 2 µmol/l; lane 3, 4 µmol/l; and lane 4, 8 µmol/l. (B) Quantification of the altered expression level. *P<0.05 vs. 0 µmol/l.
Discussion
Histone methylation is an important regulatory mechanism in changing chromatin structure and transcription, which is associated with the generation and development of different types of tumor. H3K9 monomethylation, dimethylation and trimethylation indicate transcriptional inhibition. The trimethylation of H3K9 was associated with the closed structure of heterochromatin, while H3K9 dimethylation was associated with autosomal silencing (6). Stewart et al (7) demonstrated that H3K9 downregulated acetylation of H3 and H4, and deacetylated H3 and H4, which inhibited transcription and induced gene silencing. However, in endotoxin-tolerant THP-1 cells, G9a bound to the promoter region of TNFα, to promote dimethylation of H3K9 and enriched DNMTa/3b, which promoted DNA methylation and inhibited TNFα transcription (8). H3K9 hypermethylation in the promoter region of the suppressor gene led to suppressor gene silencing, which is associated with the generation of tumors. Following treatment with 5-Aza-Dc in different types of gastric cancer cells, the dimethylation of H3K9 decreased, DNA was demethylated and P16 expression was induced (9).
A previous study demonstrated that in gastric carcinoma, histone methyltransferase Suv39H1 and H3K9 trimethylation were highly expressed (10). Further studies demonstrated that Suv39H1 and H3K9 methylation were associated with tumor differentiation, invasive depth and lymph node metastasis, which indicated that the two may possibly promote the generation, development, invasion and metastasis of gastric carcinoma (11,12). In the present study, the positive rate of H3K9me2 in glioma tissue was 82% (41/50), which was significantly different to the 38% (19/50) in adjacent tissue (χ2=18.38, P<0.01). The positive rate of H3K9me2 was associated with higher WHO glioma grade. However, the positive rate of H3K9me1 in glioma tissue was 54% (27/50), and 44% (22/50) in adjacent tissue (χ2=1.21, P>0.05). These results indicated that H3K9me2 was associated with the generation and development of glioma, while H3K9me1 was not important in the generation and development of glioma. H3K9me2 may be an indicator of glioma severity and prognosis.
Lysine methylation of histone was completed by the catalysis of histone lysine methyltransferases, while G9a is the predominant methyltransferase of H3K9. In euchromatin, G9a catalyzed monomethylation and dimethylation of H3K9, while methylated H3K27 in vitro (13). G9a gene deletion led to a decrease in H3K9me2 in the nuclear peripheral region, while some control of gene silencing was lost (14). Previous studies have demonstrated that the abnormal expression of histone methyltransferase and histone demethylase were associated with the generation and development of tumors (15). Chen et al (16) observed that the mRNA and protein expression levels of histone methyltransferase G9 gene in extrahepatic bile duct carcinoma were increased compared with the control group, which were positively associated with lymph node metastasis and TNM staging (16). The present study determined that the positive rate of G9a in glioma tissues was 86% (43/50), which was significantly different from 42% (21/50) in adjacent tissues (P<0.01). Increased H3K9me2 expression was associated with higher WHO grade. These results indicated that G9a was expressed at higher levels in tumor tissue samples than in normal tissue samples, which may be targeted in therapeutic strategies. Wu et al (9) used depsipeptide to downregulate G9a and SUV39H1 expression, leading to a decrease in H3K9me2/3 in the promoter region of P16. The enrichment of DNMT1 decreased, DNA was demethylated, P16 was expressed and the proliferation of tumor cells was inhibited.
BIX-01294 is a selective artificial inhibitor of G9a. Kondo et al (3) demonstrated that following BIX-01294 treatment, the activity of G9a and H3K9 methylation decreased, leading to P16 and RASSF1A expression and marked inhibition of PC3 prostate cancer cell growth (3). In the present study, following treatment with different concentrations of BIX-01294 (1, 2, 4 and, 8 µmol/l) for 24 h, the proliferation rates of U251 were 94.12±3.41, 78.83±2.25, 53.68±2.54 and 21.04±2.07%, respectively, which indicated there was a significant concentration-dependent increase. As the treatment time increased, the proliferation rate decreased. Following treatment with different concentrations of BIX-01294 (1, 2, 4 and 8 µmol/l) for 24 h, the apoptosis rates of U251 were 3.67±1.42, 16.42±5.18, 35.18±3.26 and 57.52±4.37%, which demonstrated the significant increase in apoptosis in a concentration-dependent manner.
Apoptosis is the spontaneous process of programmed cell death, whose generation is under rigorous control in the body. The expression and regulation of the Bcl-2 family is important in signal transduction pathways and apoptosis. The Bcl-2 family includes two types of protein, those that are anti-apoptotic, including Bcl-2, B-cell lymphoma-extra large, Bcl-2-like protein 2, myeloid cell leukemia-1 and Bcl-2-related protein A1 and pro-apoptotic proteins, including Bax, Bcl-2 homologous antagonist/killer, BH3 interacting-domain death agonist and Bcl-2-associated death promoter (17). Bcl-2 and Bax are the representative anti-apoptotic and pro-apoptotic gene in Bcl-2 family, respectively. Bcl-2 inhibits apoptosis of tumor cells induced by different factors. Bax forms an apoptosis complex and activates caspase-9, which subsequently activates the downstream effector proteins, including caspase-3, caspase-6 and caspase-7 (18). It was observed that following BIX-01294 treatment, Bcl-2 protein expression was downregulated, while Bax expression was upregulated. Apoptosis-associated proteins caspase-9 and caspase-3 were upregulated, which resulted in cell apoptosis.
G9a is able to monomethylate and dimethylate H3K9, while trimethylation is reduced when incubated in vitro for an extended amount of time. G9a and GLP in mouse euchromatin is dependent on the heterodimer formed by the SET domain, which may catalyze the production of H3K9me2 and H3K9me1, enrich heterochromatin protein 1 in euchromatin and mediate transcriptional repression (19). Knock-out of G9a in mice reduced H3K9me2 which led to the silencing of 167 genes that have enrichment of H3K9me2 on the promoter (14). The present study observed that following BIX-01294-induced inhibition of G9a activity, methylation of H3K9me1 and H3K9me2 was downregulated, however, the expression of H3K9me3 and Act-H3 is less altered, which is consistent with monomethylation and dimethylation and lacking trimethylation.
H3K27me3 was considered to be associated with transcriptional inhibition. Following trimethylation of H3K27, the protein regulator of cytokinesis 1 complex combined with the specific gene locus, then inhibited the enrichment of transcription activators, silenced tumor suppressor genes and led to carcinoma. Enhancer of zeste homolog 2, the histone methyltransferase catalyzing H3K27 trimethylation, is expressed in numerous tumors, including glioma, which was associated with development and poor prognosis (20,21). However, less research has been conducted on H3K27me1 and H3K27me2. Previous studies demonstrated that in the embryonic stem cell line with deletion of G9a, the methylation of the 27th lysine in H3 decreased and polycomb repressive complex 2 has an important role in this process (22,23). It was observed that following BIX-01294 treatment to inhibit G9a activity, the methylation of H3K27me1 and H3K27me2 decreased significantly, which suggested that G9a had the ability to methylate H3K27, however, this remains to be investigated in a clinical setting.
In conclusion, the present study observed that BIX-01294 inhibits the proliferation of glioma cells and induces apoptosis. Further study demonstrated that the alteration of methylation of H3K9 and H3K27 resulted in changes in chromosomal conformation, which may be an underlying mechanism of inhibition of tumor cells proliferation. BIX-01294 may be a potential novel therapeutic agent in the treatment of glioma.
Figure 2.
Expression of H3K9me2 in glioma and adjacent tissues (Olympus BX41; magnification, ×200). (A) Positive H3K9me2 in adjacent tissues. (B) Positive H3K9me2 in grade II glioma. (C) Positive H3K9me2 in grade III glioma. (D) Positive H3K9me2 in grade IV glioma.
Acknowledgements
This work was supported by the Natural Science Foundation of Fujian Province (grant no. 2016J01484) and the Introductive Major Project of Science Research Foundation of Fujian Province (grant no. 201212004).
References
- 1.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang J, Dorsey J, Chuikov S, Pérez-Burgos L, Zhang X, Jenuwein T, Reinberg D, Berger SL. G9a and Glp methylate lysine 373 in the tumor suppressor p53. J Biol Chem. 2010;285:9636–9641. doi: 10.1074/jbc.M109.062588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kondo Y, Shen L, Ahmed S, Boumber Y, Sekido Y, Haddad BR, Issa JP. Downregulation of histone H3 lysine 9 methyltransferase G9a induces centrosome disruption and chromosome instability in cancer cells. PLoS One. 2008;3:e2037. doi: 10.1371/journal.pone.0002037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fuller GN, Scheithauer BW. The 2007 Revised World Health Organization (WHO) Classification of Tumours of the Central Nervous System: newly codified entities. Brain Pathol. 2007;17:304–307. doi: 10.1111/j.1750-3639.2007.00084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin X, Huang Y, Zou Y, Chen X, Ma X. Depletion of G9a gene induces cell apoptosis in human gastric carcinoma. Oncology Reports. 2016;35:3041–3049. doi: 10.3892/or.2016.4692. [DOI] [PubMed] [Google Scholar]
- 6.McGarvey KM, Fahrner JA, Greene E, Martens J, Jenuwein T, Baylin SB. Silenced toumor stressor genes reactivated by DNA demethylation do not return to a fully euchromatic chromati state. Cancer Res. 2006;66:3541–3549. doi: 10.1158/0008-5472.CAN-05-2481. [DOI] [PubMed] [Google Scholar]
- 7.Stewart MD, Li J, Wong J. Elationship between histone H3 lysine 9 methylation, transcription repression, and heterochromatin protein 1 recruitment. Mol Cell Biol. 2005;25:2525–2538. doi: 10.1128/MCB.25.7.2525-2538.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El Gazzar M, Yoza BK, Chen X, Hu J, Hawkins GA, McCall CE. G9a and HP1 couple histone and DNA methylation to TNFalpha transcription silencing during endotoxin tolerance. J Biol Chem. 2008;283:32198–32208. doi: 10.1074/jbc.M803446200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu LP, Wang X, Li L, Zhao Y, Lu S, Yu Y, Zhou W, Liu X, Yang J, Zheng Z, et al. Histone deacetylase inhibitor depsipeptide activates silenced genes through decreasing both CpG and H3K9 methylation on the promoter. Mol Cell Biol. 2008;28:3219–3235. doi: 10.1128/MCB.01516-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai L, Ma X, Huang Y, Zou Y, Chen X. Aberrant histone methylation and the effect of Suv39H1 siRNA on gastric carcinoma. Oncol Rep. 2014;31:2593–2600. doi: 10.3892/or.2014.3135. [DOI] [PubMed] [Google Scholar]
- 11.Cattaneo F, Nucifora G. EVI1 recruits the histone methyltransferase SUV39H1 for transcription repression. J Cell Biochem. 2008;105:344–352. doi: 10.1002/jcb.21869. [DOI] [PubMed] [Google Scholar]
- 12.Goyama S, Nitta E, Yoshino T, Kako S, Watanabe-Okochi N, Shimabe M, Imai Y, Takahashi K, Kurokawa M. EVI-1 interacts with histone methyl transferases SUV39H1 and G9a for transcriptional repression and bone marrow immortalization. Leukemia. 2010;24:81–88. doi: 10.1038/leu.2009.202. [DOI] [PubMed] [Google Scholar]
- 13.Wu H, Chen X, Xiong J, Li Y, Li H, Ding X, Liu S, Chen S, Gao S, Zhu B. Histone methyltransferase G9a contributes to H3K27 methylation in vivo. Cell Res. 2011;21:365–367. doi: 10.1038/cr.2010.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yokochi T, Poduch K, Ryba T, Lu J, Hiratani I, Tachibana M, Shinkai Y, Gilbert DM. G9a selectively represses a class of late-replicating genes at the nuclear periphery. Proc Natl Acad Sci USA. 2009;106:19363–19368. doi: 10.1073/pnas.0906142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zou Y, Ma X, Huang Y, Hong L, Chiao JW. Effect of phenylhexyl isothiocyanate on aberrant histone H3 methylation in primary human acute leukaemia. J Hematol Oncol. 2012;5:36. doi: 10.1186/1756-8722-5-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Y, Luo J, Chen B, Wang J, Zou S. Expression of histone methyltransferase G9a and clinical significance in extrahepatic cholangiocarcinoma. Chin-Ger J Clin Oncol. 2008;7:10–13. doi: 10.1007/s10330-007-0126-0. [DOI] [Google Scholar]
- 17.Sitailo LA, Jerome-Morais A, Denning MF. Mcl-1 functions as major epidermal survival protein required for proper keratinocyte differentiation. J Invest Dermatol. 2009;129:1351–1360. doi: 10.1038/jid.2008.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Theofilas P, Bedner P, Hüttmann K, Theis M, Steinhäuser C, Frank S. The proapoptotic BCL-2 homology domain 3-only protein Bim is not critical for acute excitotoxic cell death. J Neuropathol Exp Neurol. 2009;68:102–110. doi: 10.1097/NEN.0b013e31819385fd. [DOI] [PubMed] [Google Scholar]
- 19.Tachibana M, Matsumura Y, Fukuda M, Kimura H, Shinkai Y. G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription. EMBO J. 2008;27:2681–2690. doi: 10.1038/emboj.2008.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang YB, Niu HT, Chang JW, Dong GL, Ma XB. EZH2 silencing by RNA interference inhibits proliferation in bladder cancer cell lines. Eur J Cancer Care (Engl) 2011;20:106–112. doi: 10.1111/j.1365-2354.2009.01148.x. [DOI] [PubMed] [Google Scholar]
- 21.Bryant RJ, Winder SJ, Cross SS, Hamdy FC, Cunliffe VT. The Polycomb group protein EZH2 regulates actin polymerization in human prostate cancer cells. Prostate. 2008;68:255–263. doi: 10.1002/pros.20705. [DOI] [PubMed] [Google Scholar]
- 22.Ma DK, Chiang CH, Ponnusamy K, Ming GL, Song H. G9a and Jhdm2a regulate embryonic stem cell fusion-induced reprogramming of adult neural stem cells. Stem Cells. 2008;26:2131–2141. doi: 10.1634/stemcells.2008-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ikegami K, Iwatani M, Suzuki M, Tachibana M, Shinkai Y, Tanaka S, Greally JM, Yagi S, Hattori N, Shiota K. Genome-wide and locus-specific DNA hypomethylation in G9a deficient mouse embryonic stem cells. Genes Cells. 2007;12:1–11. doi: 10.1111/j.1365-2443.2006.01029.x. [DOI] [PubMed] [Google Scholar]