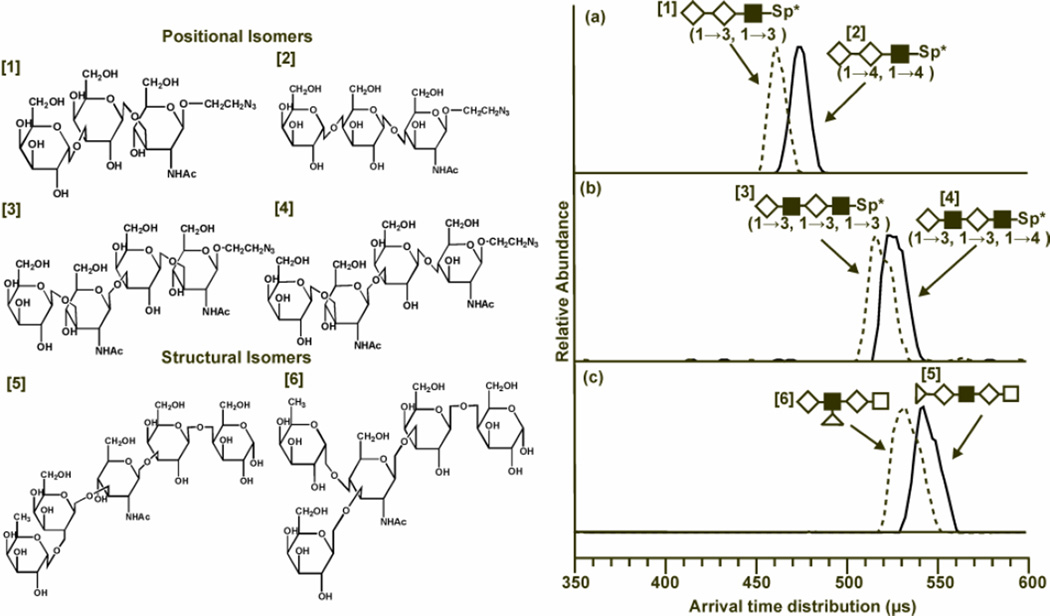
Figure 3.
Structures of the isobaric sets of positional and structural isomers (left) and the associated drift time profiles (right). (Left) Note the difference in structures between glycans 1 and 2 are two 1→3 glycosidic linkages being replaced with two 1→4 linkages. Glycans 3 and 4 have one linkage variation, and glycans 5 and 6 (LNFP1 and LNFP2) vary in the location of fucose from galactose to N-acetylglucoseamine. (Right) Drift time profiles at an electrostatic field strength of 20.6 volts cm−1 Torr within the ion mobility drift cell. Structures of the oligosaccharides are replaced with shape representations. Drift times are related to the ion structure in that larger, more elongated ions experience more collisions with the neutral buffer gas present in the drift cell causing a longer drift time than more compact structures. (a) In the comparison between glycans 1 (dotted line) and 2 (solid line), the 1→3 linkages of glycan 1 cause it to have a shorter drift time which indicates a more compact structure that glycan 2, which is more elongated. (b) Glycans 3 (dotted line) and 4 (solid line) have differing drift times due to the change in one glycosidic linkage. The 1→3 linkages allow glycan 3 to adopt a more compact conformation when compared to its positional isomer, which has one 1→4 linkage. (c) Drift time profiles for glycans 5 (solid line) and 6 (dotted line) are compared. LNFP2 has a shorter drift time than LNFP1 at both voltages. This is attributed to increased branching in LNFP2 that allows the glycan to adopt a more compact structure. Individual monosaccharide representations are as follows: ◊ - galactose; ■-N-acetylglucoseamine; Δ -fucose; with linkage information for the positional isomers provided in the parenthesis below each representation (54). Fenn, L. S., and McLean, J. A. (2011) Structural resolution of carbohydrate positional and structural isomers based on gas-phase ion mobility-mass spectrometry. Phys Chem Chem Phys 13, 2196–2205- Reproduced by permission of the PCCP Owner Societies.