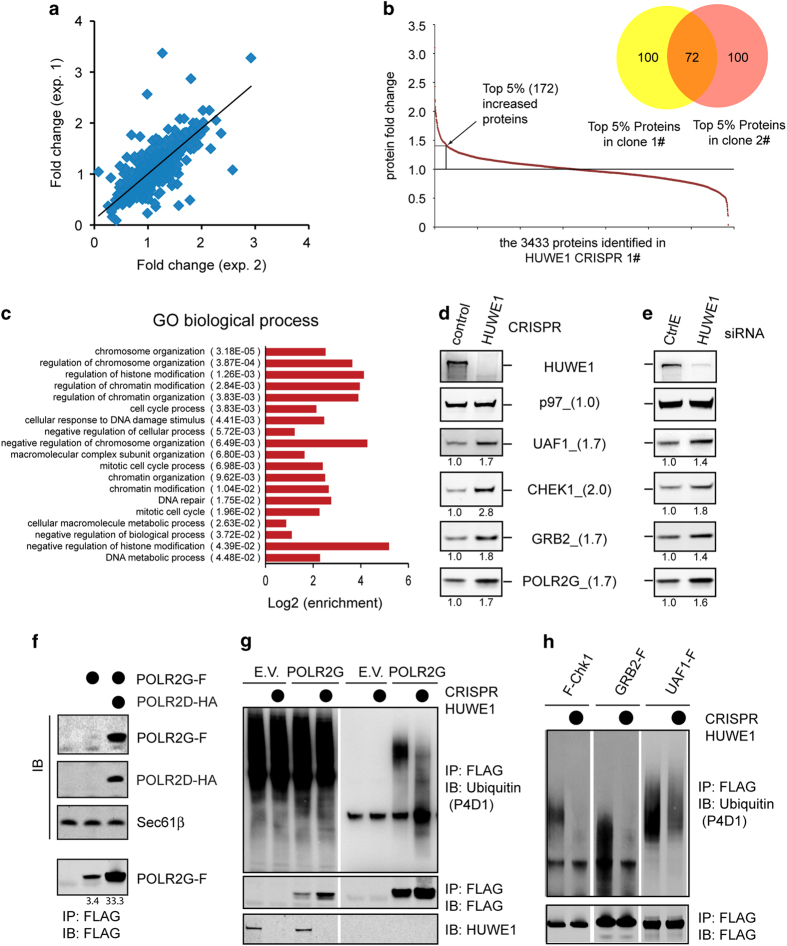
Figure 5.
Identification of endogenous HUWE1 substrates. (a) Protein fold change comparison between two independent SILAC experiments (exp.). (b) The range of protein fold changes for the 3 433 proteins identified in HUWE1 knockout clone 1. The Venn diagram compared the top 172 proteins increased upon HUWE1 inactivation in the two different CRISPR clones. (c) The biological processes affected by HUWE1 inactivation. (d) Immunoblotting analyses validate the stabilization of proteins involved in DNA damage response pathways using HUWE1 CRISPR cells. The numbers in parentheses indicate fold changes measured by SILAC. The numbers under each gel panel show the band intensity. (e) Immunoblotting validation using HEK293T cells transfected with control or HUWE1 siRNA. (f) Unassembled POLR2G is unstable. Immunoblotting analysis of cells transfected with FLAG-tagged POLR2G (POLR2G-F) either alone or together with HA-tagged POLR2D. (g, h) The ubiquitination state of substrates in control and HUWE1 CRISPR cells. (g) Control or HUWE1 CRISPR cells were transfected with either an empty vector (EV) or a construct expressing FLAG-tagged POLR2G. Cell lysate were subject to immunoprecipitation under denaturing conditions and analyzed by immunoblotting. (h) Cell lysates from control or HUWE1 CRISPR cells transfected with the indicated HUWE1 substrates were subject to immunoprecipitation using FLAG beads. Precipitated proteins were analyzed by immunoblotting.