Extended Data Figure 2. Consistency among individual STP tomography image sets.
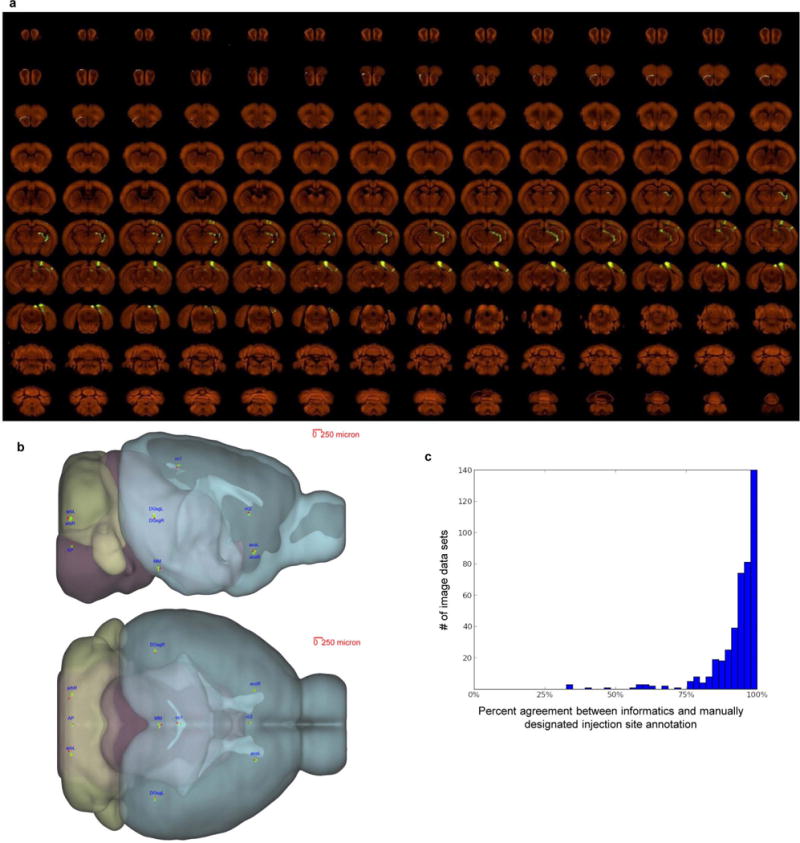
a, High-resolution images of 140 serial sections of a single brain are shown as an example (injection into the primary visual area). The injection site and major projection targets can be easily observed in this ‘contact-sheet’ view. b, Registration variability study. A set of ten 3D fiducial points of interest (POIs) were manually identified on the Nissl 3D reference space, the average template brain, and in 30 randomly selected individual image sets by three raters independently. The POIs were selected such that they span the brain and can be easily and repeatedly identified in 3D. Each POI (p) from each experiment was then projected into 3D reference space (p’) using the transform parameters computed by the Alignment module. Statistics were gathered on the target registration error between p’ and its ‘gold standard’ correspondence (computed as the mean of the labelling of 3 raters) in the Nissl (pNissl) and template (ptemplate) volumes. In summary, for all POIs in template space, the observed median variation in each direction are 28 μm for left-right, 35 μm for inferior-superior, and 49 μm for anterior-posterior. In Nissl space, these observed median variations are 42, 71 and 60 μm, respectively. The registration variations in different directions are shown here. For each POI, the green dot shows the position in template space (ptemplate) and the red dot shows the position in Nissl space (pNissl). The small yellow bar shows the median variation (among 30 image sets) away from ptemplate in each direction. POIs: AP: area postrema, midline; MM: medial mammillary nucleus, midline; cc1: corpus callosum, midline; cc2: corpus callosum, midline; acoL, acoR: anterior commisure, olfactory limb; arbL, arbR: arbor vitae; DGsgL, DGsgR: dentate gyrus, granule cell layer. c, Percent agreement between computationally assigned injection site voxels and manually assigned injection structures. Each histogram data point corresponds to a single injection, and 100% indicates that every injection site voxel was computationally assigned to a structure included on the manually annotated injection structures list. Voxels which were computationally assigned to fibre tracts or ventricles are excluded from the computation. Neither fibre tracts nor ventricles were incorporated into the manual annotation process; their exclusion allows for a more commensurate comparison.
