Abstract
Module-based network analysis of diverse pharmacological mechanisms is critical to systematically understand combination therapies and disease outcomes. We first constructed drug-target ischemic networks in baicalin, jasminoidin, ursodeoxycholic acid, and their combinations baicalin and jasminoidin as well as jasminoidin and ursodeoxycholic acid groups and identified modules using the entropy-based clustering algorithm. The modules 11, 7, 4, 8 and 3 were identified as baicalin, jasminoidin, ursodeoxycholic acid, baicalin and jasminoidin and jasminoidin and ursodeoxycholic acid-emerged responsive modules, while 12, 8, 15, 17 and 9 were identified as disappeared responsive modules based on variation of topological similarity, respectively. No overlapping differential biological processes were enriched between baicalin and jasminoidin and jasminoidin and ursodeoxycholic acid pure emerged responsive modules, but two were enriched by their co-disappeared responsive modules including nucleotide-excision repair and epithelial structure maintenance. We found an additive effect of baicalin and jasminoidin in a divergent pattern and a synergistic effect of jasminoidin and ursodeoxycholic acid in a convergent pattern on “central hit strategy” of regulating inflammation against cerebral ischemia. The proposed module-based approach may provide us a holistic view to understand multiple pharmacological mechanisms associated with differential phenotypes from the standpoint of modular pharmacology.
Keywords: Modular pharmacology, emerged responsive module, disappeared responsive module, additive effect, synergistic effect, cerebral ischemia
Introduction
Since the advent of high-throughput experimental techniques, the complexity of biological networks has become increasingly evident,1 and it has also been shown that individual genes or proteins are often difficult to be placed in biological context and fully annotate functionally.2 Concomitantly, the study of pharmacological mechanisms in treatment of complex diseases such as cerebral ischemia is re-addressed using systems pharmacology approaches and systems medicine.3,4 As such, requirement for “network design principles” became both feasible and necessary. During the last decade, the modular nature of a wide variety of complex networks has been investigated in detail,5–7 and modules as the building blocks of higher level functional organization have played a key role in decoding the mechanism of complex systems.8 Moreover, use of modular design as a tool in pharmacological research contributes a lot to rationalize drug actions and predict disease outcomes.9,10 For example, modular biomarkers in response to tamoxifen attest to the immunomodulatory role of tamoxifen, and further reveal that it deregulates cell cycle and apoptosis pathways.11 A module network rewiring-analysis predicts dynamical drug sensitivity and resistance, and also characterizes complex dynamic processes for therapy response.12 Characterization of drug-induced transcriptional modules has provided a starting point for drug repositioning and functional understanding.13 Therefore, the study of therapy response basing on modules, cannot merely elucidate essential principles to react to drug at the network level, but also reveal fundamental mechanisms, thereby benefitting efficient identification of biomarkers in pharmacogenomics.14,15
Qingkailing injection, an extensively used traditional Chinese patent medicine in treating cerebral ischemia in China, can reduce intracerebral hemorrhage-induced brain damage as well as ischemic stroke-induced infarct volume through inhibiting apoptosis and increasing the expression of endothelial nitric oxide synthase.16 Baicalin (BA), jasminoidin (JA), and ursodeoxycholic acid (UA) are the three bioactive ingredients of Qingkailing injection.17 The pharmacological mechanisms of these three ingredients and their combinations including BA and JA (BJ) as well as JA and UA (JU) in the treatment of cerebral ischemia have shown both similarities and differences according to analysis of differentially expressed genes and signaling pathways. BA, UA and JA all act on Ca2+-dependent signaling cascades.18 BA activates extracellular matrix-receptor interaction pathways, leading to direct or indirect control of cellular activities.19 JA inhibits the expression of caspase-3 after brain damage.20 UA results in the downregulation of Bdnf, MNK1/2 (Mknk1), and c-fos (Fos), regulating cell proliferation and differentiation.19 Besides, JU plays an important role of the synergistic effect in activating P53 pathway,21 while BA and JA fuse those differentially expressed genes to contribute to the additive effect of BJ, such as Huntington’s disease signaling.22
In this context, moving a step further, we developed a novel module-based method to identify drug responsive modules by integrating gene expression data and high-quality protein–protein interaction (PPI) networks. Our goal was to demonstrate that the response mechanisms of different compound treatments on cerebral ischemia could be elucidated from a modular pharmacology standpoint9 and predict drug targets in a holistic view. Specifically, using this method, we compared the pharmacological mechanisms of the two combination therapies BJ and JU in treating cerebral ischemia.
Materials and methods
Data preparation
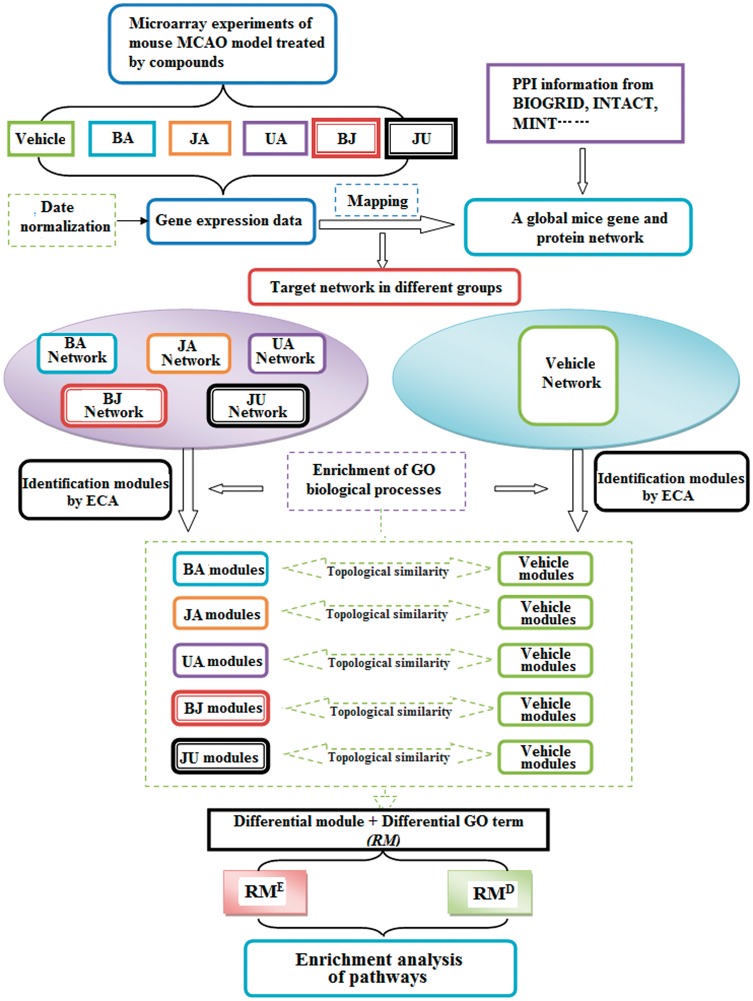
The gene expression data were obtained from our previous study, and six group datasets were included: vehicle, BA, JA, UA as well as two combinations BJ and JU.23 A microarray chip containing 16,463 oligoclones (Incyte Genomics, Santa Clara, CA, USA) was used to conduct gene expression profiling. The procedures of microarray preparation and data analysis have been described previously.21–23 The experimental protocol and data analysis are shown in Figure 1.
Figure 1.
Flow diagram. Drug-target networks were constructed by integrating gene expression and protein interaction data, and the modules were identified using entropy-based clustering algorithm (ECA). The comparison of topological similarity and GO functional enrichment analysis was used to define the drug responsive modules (RMs). The KEGG pathway in RMs was enriched using DAVID 6.7 software. (A color version of this figure is available in the online journal.)
Constructing target networks in different groups
We downloaded PPI data from NIA Mouse Protein–Protein Interaction Database,24 INTACT,25 BIOGRID,26 and MINT,27 and then deleted duplicate data and self-interactions. In our analysis, we not only considered interactions present in these databases but also predicted orthologous and paralogous data from the mouse PPIs. We constructed a unique global mouse gene and protein network by integrating protein interactions.
We used our previous gene expression profiles of hippocampus in ischemic mice treated with the three monotherapies (BA, UA, and JA) and the two combinations (BJ and JU). The mean centered normalization of expressed data was performed. Genes with normalized expression value greater than one (that is, the original expression value is greater than the mean) were defined as significantly differentially expressed. Finally, we generated target networks of each group by mapping these genes to the interaction network.
A plugin for Cytoscape Network Analyzer28 was employed to compute and display a comprehensive set of topological parameters, including the network nodes, edges, density, clustering coefficient, connected components, diameter, radius, and centralization.
Identifying modules in different groups
We selected the entropy-based clustering algorithm (ECA)29 to mine modules in the related target networks of each group. This algorithm exploits the concept of entropy to assess modularity of a graph and defines loss of entropy representing an increase in modularity.29 Since low entropy indicates low uncertainty which represents a stable state,30,31 we expect this algorithm could find a stable modular state. It first selects a random seed as the initial node and forms a seed cluster by including all neighbors of the seed. Next, the seed cluster shrinks and grows to minimize graph entropy by iteratively removing and adding the nodes on the borders of the cluster. This seed growth stops when an optimal local boundary is found. The process of selecting a seed and generating an optimal cluster is reiterated, leading to a set of clusters. We set the graph entropy less than a threshold of 20 as the cutoff value to filter out clusters and 3 as the minimum module size. To eliminate the randomness, we subsequently identified modules randomly in all the six networks and then compared the average graph entropy with the ECA result.
Gene ontology enrichment analysis
Based on the detected modules, gene ontology (GO) enrichment analysis was utilized to characterize the function of modules by using the GO::Term Finder tool (http://search.cpan.org/dist/GO-TermFinder/)32–34 and mouse genome informatics.35 An over representation of a term is defined as a modified Fisher’s exact p-value with an adjustment for multiple tests using Benjamini method. In this analysis, all the genes on the array were set as the background, and GO terms with P value <0.05 were considered as significant.
Defining the drug responsive module (RM), based on topological similarity
To define the RM, we first quantified the topological similarity between modules in vehicle and any treatment groups (BA, JA, UA, BJ or JU group). We measured the similarity between modules G1 and G2 in the vehicle and any treatment groups
The term () denotes the number of common edges and () represents the number of edges in the union of G1 and G2. The similarity ranges from 0 to 1 and we set a threshold of 0.9 as the cutoff. Similar modules were defined in terms of the value of similarity greater than 0.9. If the similarity of one module compared with any module in the compared group was less than 0.9, we defined it as a differential module. In other words, it is a module that was absent in the compared groups. We also integrated the GO enrichment analysis: if a GO term has just been enriched in one differential module compared with modules in other groups, we defined it as a differential function of this differential module and the module was accordingly a drug responsive module (RM). We further divided it into an emerged RM (RME) or a disappeared RM (RMD). For example, if an RM in the vehicle group was not identified in the treated groups, meaning that the module vanished after drug intervention, we defined it as an RMD. On the other hand, if an RM just appeared in the treated groups, we considered it as an RME.
RMs pathway analysis
Enrichment analysis of KEGG pathways on RMs was performed using a web server DAVID 6.7 functional annotation tool (http://david.abcc.ncifcrf.gov/).36 We selected all pathways with a P value <0.05 after correcting for multiple terms, tested by Benjamini Notably, we only enriched the pathways that corresponded to the differential functions.
Results
Pharmacodynamic results
It was found that BA, JA and UA effectively reduced the ischemic infarct volume compared with the vehicle group (P <0.05).17 BJ was more effective than BA or JA monotherapy in reducing the ischemic infarction volume, and similar result was obtained by JU compared with JA or UA alone.23 Further analysis demonstrated that BJ showed additive effects,22 and JU resulted in synergistic effects.21,37
Network topological parameters in different groups
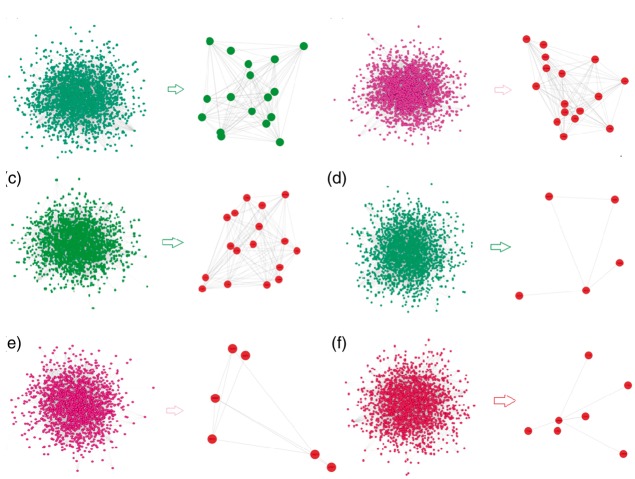
The experimental protocol and data analysis are shown in Figure 1.We generated a mouse protein interaction network containing 65,850 edges by integrating multiple protein interaction databases. After mapping the different experimental gene expression data to this work, we finally obtained five drug interventional target networks of BA, JA, UA, BJ and JU as well as one vehicle target network (Figure 2). The network topological parameters such as nodes, edges, density, clustering coefficient, connected component, diameter, radius and centralization of these six networks were similar (Table 1).
Figure 2.
Global networks and identified modules of vehicle, BA, UA, JA, BJ and JU groups. (A color version of this figure is available in the online journal.)
Table 1.
Topological attributes of global networks in different groups
| Topological attributes | Vehicle | BA | JA | UA | BJ | JU |
|---|---|---|---|---|---|---|
| Nodes | 2183 | 2229 | 2032 | 1981 | 1899 | 2028 |
| Edges | 6656 | 6738 | 5980 | 5780 | 5415 | 6108 |
| Density | 0.003 | 0.003 | 0.003 | 0.003 | 0.003 | 0.003 |
| Diameter | 12 | 11 | 12 | 13 | 12 | 11 |
| Radius | 1 | 1 | 1 | 1 | 1 | 1 |
| Centralization | 0.129 | 0.128 | 0.136 | 0.136 | 0.137 | 0.134 |
| Clustering coefficient | 0.109 | 0.102 | 0.109 | 0.106 | 0.103 | 0.106 |
| Connected components | 26 | 27 | 26 | 25 | 25 | 21 |
Module distribution
The modules identified by ECA are shown in Table 2. We identified 841, 749, 740, 690, 770 and 792 modules (nodes ≥3) from BA, JA, UA, BJ, JU and vehicle target networks, respectively. The average size of those modules ranged from 3.81 to 3.90. The values of average entropy of the six networks were calculated (range, 1.242 to 1320), which were similar to each other but significantly lower than that with the randomized identification (range, 3.10–3.31) (P<0.05).
Table 2.
Modules identified by entropy-based clustering algorithm (ECA) in different groups
| Groups | Clusters | Average size | Sum of the entropy | Average entropy | Average entropy in randomized networks |
|---|---|---|---|---|---|
| Vehicle | 792 | 3.871 | 985.87 | 1.245 | 3.280 |
| BA | 841 | 3.900 | 1094.94 | 1.302 | 3.201 |
| JA | 749 | 3.854 | 930.49 | 1.242 | 3.279 |
| UA | 740 | 3.820 | 932.86 | 1.261 | 3.203 |
| BJ | 690 | 3.896 | 910.73 | 1.320 | 3.098 |
| JU | 770 | 3.812 | 943.61 | 1.225 | 3.297 |
RMs among different monotherapies
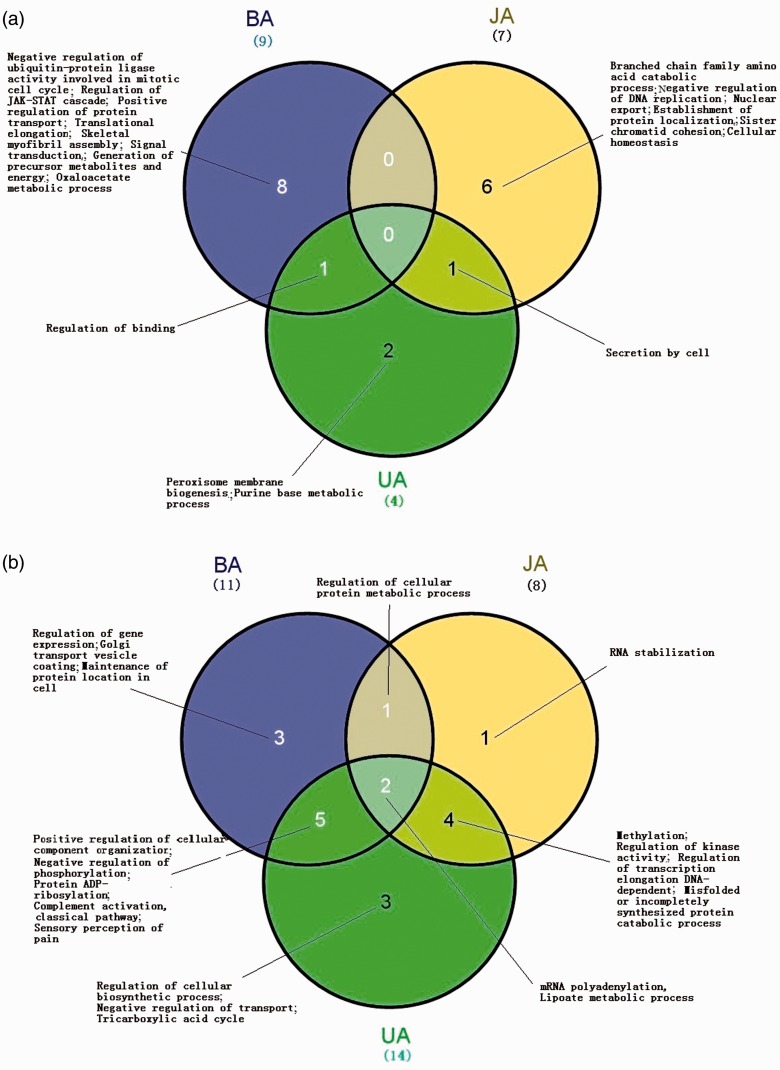
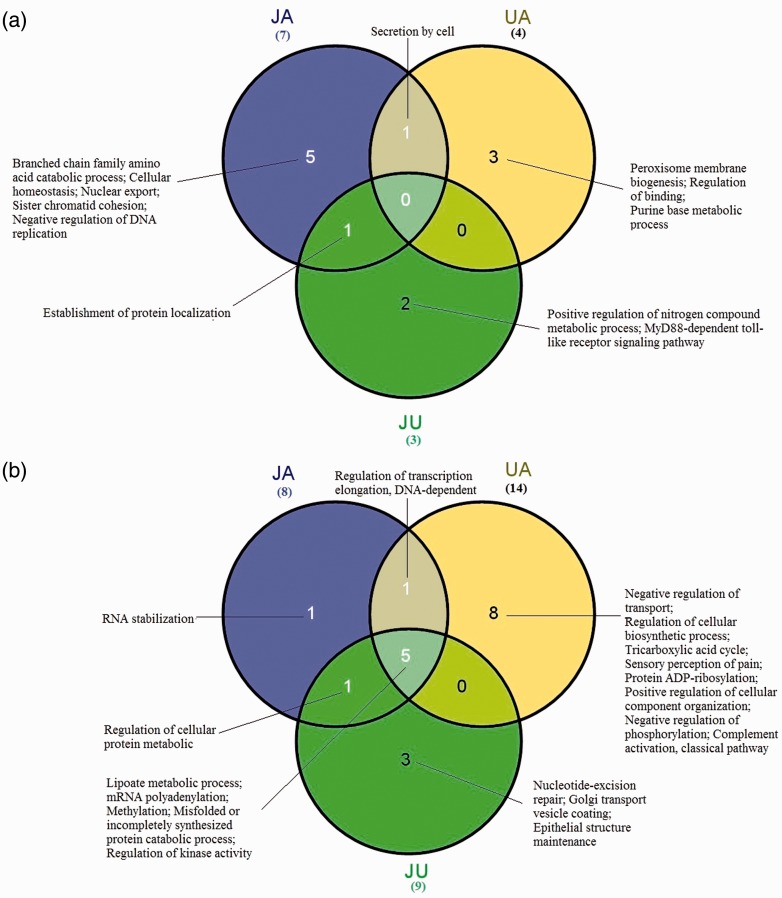
Results showed that 64, 41 and 44 differential modules were identified from BA, JA and UA groups compared with vehicle group, respectively. Integrating the GO enrichment analysis, 11, 7 and 4 modules were identified as the RMEs of BA, JA and UA, respectively. Different modules in the enrichment might correspond to the same biological processes. Finally, there were 9, 7 and 4 significantly enriched differential biological processes belonging to the relevant modules (Figure 3(a)). Regarding to the overlapping differential biological processes among the RMEs, none were found among the three, but one was observed between the BA and UA groups (regulation of binding) and another one between the BA and JA groups (secretion by cell). No overlap of differential biological processes was found between the UA and JA groups (Figure 3(a)). Furthermore, eight (88.9%), six (85.7%) and two (50%) differential biological processes were only enriched in the BA, JA and UA-RMEs, respectively.
Figure 3.
Differential biological processes enriched by RMs between BA, JA and UA. (a) Wayne map of differential biological processes enriched by RMEs between BA, JA and UA; different RMEs in the enrichment may correspond to the same biological processes, the GO term “negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle” was enriched by three different RMEs of BA. (b) Wayne map of differential biological processes enriched by RMDs between BA, JA and UA; different RMDs in the enrichment may correspond to the same biological processes, the overlap GO term “sensory perception of pain” both in BA and UA was enriched by two different RMDs, respectively. (A color version of this figure is available in the online journal.)
The RMDs were defined as modules that were present in the vehicle group but not identified in the compound-treated groups according to the similarity. We found 54, 45 and 53 differential modules in the vehicle group compared with the BA, JA and UA groups, respectively, and finally identified 12, 8 and 15 modules as BA, JA and UA-RMDs by integrating the GO enrichment analysis. Excluding the duplicates of biological processes in different modules, we obtained 11, 8 and 14 significantly enriched differential biological processes corresponding to the relevant RMDs (Figure 3(b)). Among these, two overlapping differential biological processes were identified among the BA, JA and UA groups, including mRNA polyadenylation and lipoate metabolic process. Another five overlapping differential biological processes were observed between BA and UA, 1 between BA and JA, and 4 between JA and UA, respectively. In addition, three (27.3%) differential biological processes were only enriched in the BA-RMDs, including regulation of gene expression, golgi transport vesicle coating, and maintenance of protein location in cell; three (21.4%) were only found in the UA-RMDs, including regulation of cellular biosynthetic process, negative regulation of transport, and tricarboxylic acid cycle; and one (12.5%) about RNA stabilization was only enriched in the JA-RMDs. This indicated that these compounds induced more co-disappeared modules than emerged modules.
Most of the enriched GO functions were reported to have a specific relationship with cerebral ischemia/stroke in previous studies (Supplementary Table S1, S2). For example, one of the BA-RMEs was annotated with regulation of Janus kinase/signal transducers and activators of transcription (JAK-STAT) cascade. JAK/STAT carrying signals from the cell membrane to the nucleus in response to extracellular growth factors and cytokines has been demonstrated in the astroglial response to focal cerebral ischemia.38 The therapeutic effect of BA might be partly mediated by the JAK/STAT pathway. Studies have shown that branched-chain amino acids are reduced in ischemic stroke, and the degree of reduction correlates with worse neurological outcomes.39 JA activated the “branched-chain family amino acid catabolic process” related modules independently. We inferred that the branched chain family amino acid catabolism might be one of the therapeutic targets of JA. As for the UA group, although the GO term of “peroxisome membrane biogenesis” has not been reported to be associated with cerebral ischemia directly, the impaired peroxisomal function showed significant contribution to the prolongation and intensification of inflammation.40 In contrast, UA plays a critical anti-inflammatory role based on previously reported pharmacological mechanisms in cerebral ischemia.19
RMs variation in additive effects
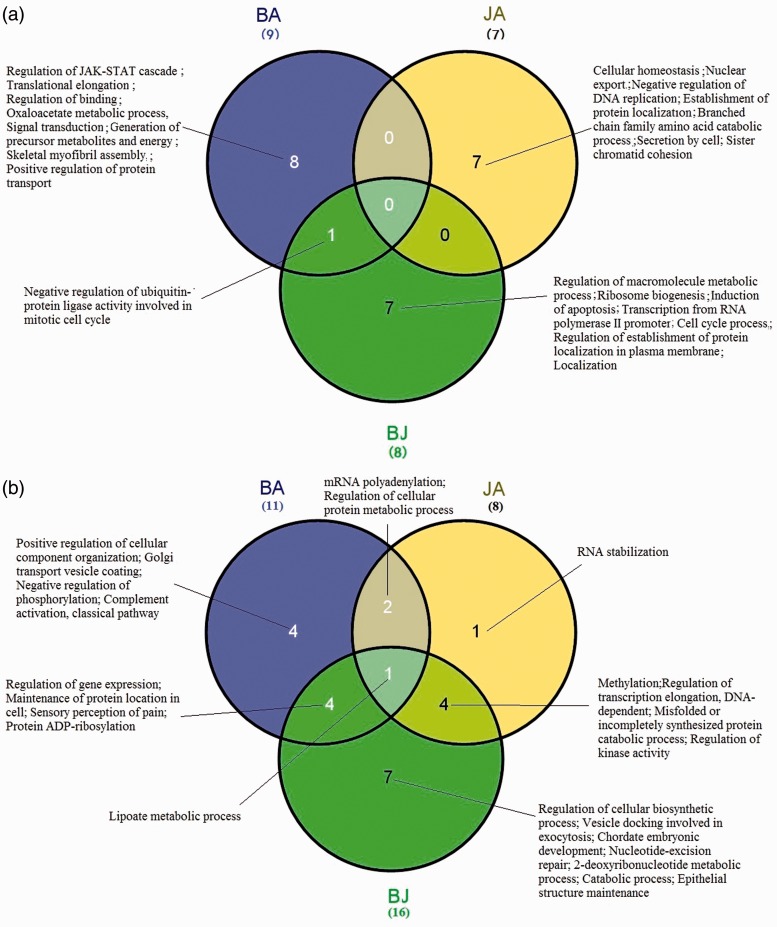
Previous studies have shown that the combination group BJ was more effective than BA or JA alone in reducing the ischemic infarction volume, indicating the underlying additive effects.22 Our module-level analysis revealed 53 differential modules in BJ group (compared with vehicle group) and 65 differential modules in vehicle group (compared with BJ group). Of these, eight were identified as BJ-RMEs including eight significantly enriched differential biological processes and 17 were BJ-RMDs including 16 significantly enriched differential biological processes. Nevertheless, the enriched BJ functions were not simply the sum of BA and JA (Figure 4(a) and (b)). Comparison of drug-RMEs revealed no overlap in differential biological processes between BA, JA and BJ as well as between JA and BJ. However, one overlapping differential biological process—negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle—was observed between BA and BJ. Comparison of drug-RMDs revealed an overlapping differential biological process related to lipoate metabolic process between BA, JA and BJ. Another four overlaps were noted between BA and BJ as well as JA and BJ. Finally, seven independent enrichment processes were used for BJ-induced RMs (87.5% RME vs. 43.8% RMD), respectively (Figure 4(a) and (b)). Interestingly, in BJ independently induced RMs, only a few GO functional annotations (3/7 in RMEs and 2/7 in RMDs) were closely related to cerebral ischemia (Supplementary Table S3, S4). GO terms such as regulation of macromolecule metabolic process or vesicle docking involved in exocytosis were not associated with cerebral ischemia/stroke.
Figure 4.
Differential biological processes enriched by RMs between BA, JA and the combination BJ. (a) Wayne map of different biological processes enriched by RMEs between BA, JA and BJ. (b) Wayne map of different biological processes enriched by RMDs in BA, JA and BJ. The GO term “sensory perception of pain” in BJ was enriched by two different RMDs. (A color version of this figure is available in the online journal.)
RMs variation in synergistic effects
In contrast to the additive effects of BJ, the combination of JA and UA (JU) showed synergistic effects.37 In our module-level analysis, 53 differential modules from JU group (compared with vehicle group) and 57 differential modules from vehicle group (compared with JU group) were found. Despite the synergistic effect, JU was not related to more responsive modules than JA and UA (Figure 5(a) and (b)). Only three modules were identified as JU-RME and nine as JU-RMD. No overlapping differential biological processes were noted between JA, UA and JU-RMEs, although a total of five overlapping differential biological processes were identified among their RMDs, including lipoate metabolic process, mRNA polyadenylation, methylation, misfolded or incompletely synthesized protein catabolic process, and regulation of kinase activity (Figure 5(b)). JA-JU comparisons shared an overlap both in RMEs and RMDs, but no more were found between UA and JU. This finding is consistent with our previous results obtained from genes and pathway analysis showing that JA contributed more when administered as part of the combination JU.21,37 Furthermore, the number of the independently enriched JU-RME or -RMD modules was two (66.7%) and three (33.3%), respectively (Figure 5(a) and (b)). Based on published studies (Supplementary Table S3), only “MyD88-dependent toll-like receptor signaling pathway” in RME and “Nucleotide-excision repair” in RMD are directly related to cerebral ischemia (Supplementary Table S4).
Figure 5.
Differential biological processes enriched by RMs in JA, UA and the combination JU. (a) Wayne map of differential biological processes enriched by RMEs between JA, UA and JU. (b) Wayne map of differential biological processes enriched by RMDs between JA, UA and JU. (A color version of this figure is available in the online journal.)
Overlapping and special pure RMs between additive and synergistic effects
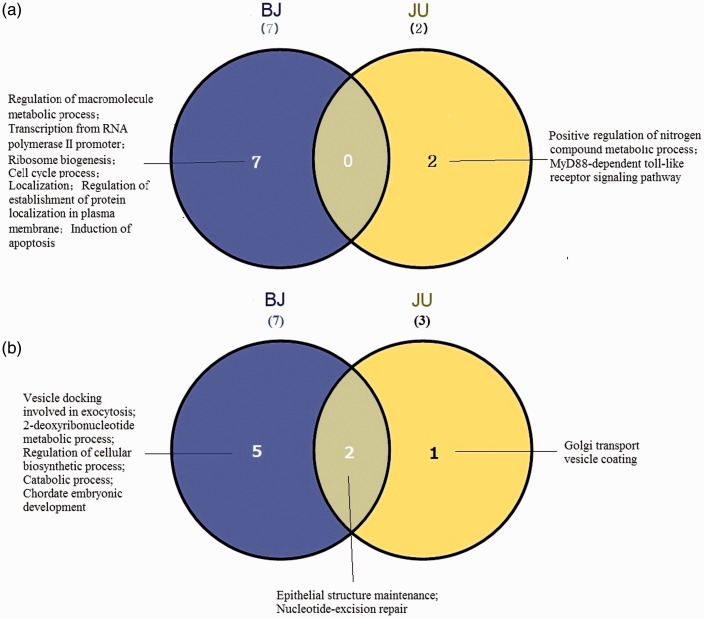
After removing the overlap with single compounds (BJ vs. BA or JA and JU vs. JA or UA), we observed both overlapping and unique pure RMs between BJ and JU (Figure 6). According to the above analysis (Figures 4 and 5), the number of RME or RMD modules with pure enrichment was both 7 for BJ, 2 and 3 for JU. Compared with BJ, JU induced fewer pure modules independently (two JU-RME vs. seven BJ-RME; one JU-RMD vs. five BJ-RMD). No overlapping differential biological processes were enriched between BJ- and JU-RMEs (Figure 6(a)), but two were enriched by their co-RMDs including nucleotide-excision repair and epithelial structure maintenance (Figure 6(b)).
Figure 6.
Differential biological processes between BJ and JU pure RMs. (a) Wayne map of different biological processes enriched by pure RMEs between BJ and JU. (b) Wayne map of different biological processes enriched by pure RMDs between BJ and JU. (A color version of this figure is available in the online journal.)
Pathway enrichment analysis of RMs
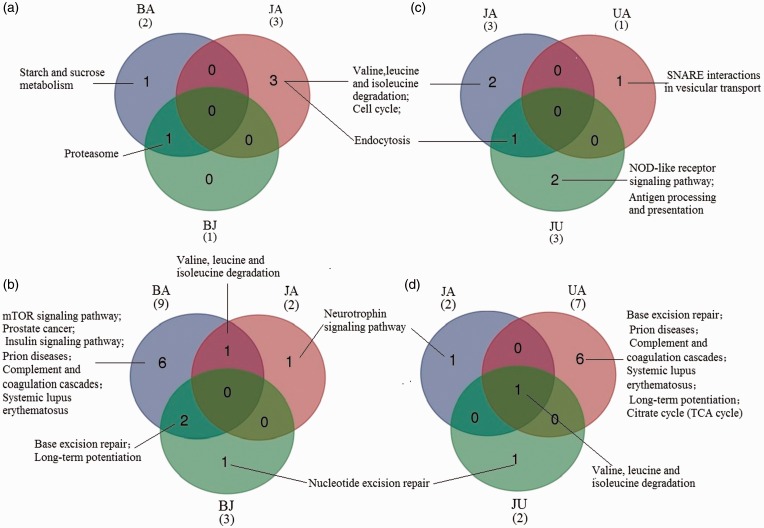
According to KEGG pathway analysis, 2, 3 and 1 pathways were significantly enriched based on the genes associated with differential biological processes corresponding to the BA, JA, and BJ-RMEs, respectively (Figure 7(a)). No overlapping pathways were noted among them, but one was found between BA and BJ-RMEs involving proteasome. For the RMDs, 9, 2 and 3 pathways were significantly enriched in the BA, JA, and BJ, respectively (Figure 7(c)). Two overlapping pathways were identified between BA and BJ as well as one between BA and JA. BA appeared to have more directional mechanism of action, inhibiting six pathways independently, including mTOR signaling pathway, prostate cancer, insulin signaling pathway, prion diseases, complement and coagulation cascades, and systemic lupus erythematosus.
Figure 7.
KEGG pathway enrichment of RMs. (a) Wayne map of pathway enrichment by RMEs between BA, JA, and BJ. (b) Wayne map of pathway enrichment by RMDs between BA, JA, and BJ. (c) Wayne map of pathway enrichment by RMEs between JA, UA and JU. (d) Wayne map of pathway enriched by RMDs between JA, UA and JU. One overlapping pathway of nucleotide excision repair was identified in BJ and JU- RMDs. (A color version of this figure is available in the online journal.)
In the JA, UA, and JU groups, KEGG pathway analysis indicated that 3, 1 and 3 pathways were significantly enriched in RMEs based on the genes corresponding to differential biological processes, respectively (Figure 7(b)). One overlapping pathway involving endocytosis was found between JA and JU, but there was no overlap among the three. For RMDs, a pathway involved in valine, leucine and isoleucine degradation was identified among the three groups, and the number of independent enrichment was 1, 6, and 1, respectively (Figure 7(d)). Based on the comparison of BJ and JU-RMEs, no overlapping pathways were found, but one overlapping pathway involved in nucleotide excision repair was identified between BJ and JU-RMDs.
The “starch and sucrose metabolism” in an RME of BA and “antigen processing and presentation” in an RMD of JU has not been reported to be associated with cerebral ischemia. However, most of the pathways have been directly or indirectly related to this disease (Supplementary Table S5, S6). Additionally, the biological significance of the enriched GO functions and pathways was not similar, and the number of the enriched GO functions was greater than that of the enriched pathways.
Discussions
Due to the complexity of diseases with multiple targets, modules are considered to be stable groups in biological networks, and may be quite robust that are not likely to be affected by individual gene expression changes.41 Therefore, a module biomarker may provide more implications to infer drug actions.42 In this paper, we developed a novel method to identify drug RMs and elucidated the potential pharmacological mechanisms of different compounds individually (BA, JA or UA) and in combinations (BJ or JU) in treating cerebral ischemia-reperfusion injury. In contrast to conventional studies on drug response that mainly focused on genes and pathways,43–45 our method focused on modules and analyzed both their topological structures and functions, which provided a holistic view of the overall differences and similarities in the pharmacological effects of the five compounds at a systems level.
All of the compounds contained more RMDs than RMEs, indicating that several modules vanished after drug intervention. At this point, it is worth emphasizing the terms of “emerged” and “disappeared”. “Emerged” refers to a group of nodes (genes or proteins) previously dissociated functionally is currently associated functionally as a module. Accordingly, “disappeared” refers to a lost module. In other words, nodes lose their co-expression relative to their interacting partners to form a structural aggregate. Though both of them may reflect the drug actions, the disappeared modules tend to load more disease-associated pharmacological mechanisms. From a modular biomarker perspective, the emergence or loss of modules reflecting functional reorganization47,48 may have pharmacological implications at the systems level and be regarded as potential therapeutic targets in cerebral ischemia. For example, all of these five compounds have one co-RMD that is annotated with GO term “lipoate metabolic process”. Lipoic acid metabolism defects are metabolic disorders that cause neurological impairment,49and lipoate confers protective effects against reperfusion injury-induced oxidative stress.50,51 In terms of pathways, valine, leucine and isoleucine degradation are other metabolic events enriched in this co-module, which closely relates to cerebral ischemia.52 This finding led us to speculate that modules carrying the above metabolic information may be regarded as useful therapeutic targets for cerebral ischemia.
Specially, another important work in our study is to explore the mechanism of combination therapies. The RMs analysis offers an alternative way to understand different mechanisms between additive and synergistic effects in fighting against cerebral ischemia.
We identified BJ and JU co-RMDs that are annotated with nucleotide-excision repair (as well as pathway enrichment) and epithelial structure maintenance, respectively. Although neither of them has been reported to be associated with cerebral ischemia directly, nucleotide-excision repair as a DNA repair mechanism may be implicated in the pathogenesis of neurologic disorders.53 Preservation of tubular epithelial structure has also been shown to prevent renal ischemia-reperfusion injury.54 Therefore, we speculate that the BJ and JU co-modules may be viewed as universal therapeutic targets for cerebral ischemia.
Among the unique BJ-induced responsive modules, divergent biological functions were enriched such as metabolism, biogenesis, apoptosis and development. Despite annotating only three RMEs and one RMD with cerebral ischemia-relevant GO terms, we hypothesize that multiple responsive modules coordinate or interact with one another to achieve a complex biological process that may contribute to the additive effect of BJ, which is also consistent with our previous study from a pathway perspective.22
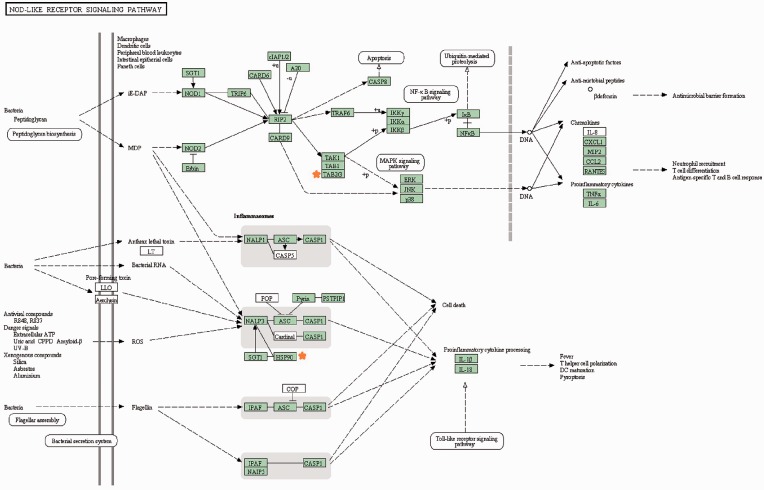
Compared with BJ, JU induced fewer unique RMs (two emerged and one disappeared). The information of RMEs enriched such as MyD88-dependent toll-like receptor signaling pathway (GO term) or NOD-like receptor signaling pathway (pathway analysis, Figure 8) have recently emerged as central regulators of immunity and inflammation-associated diseases.55,56 Increasing evidence indicates that the ischemic inflammatory response plays a detrimental role in cerebral ischemia outcome.57,58 Modulation of inflammatory signaling may be a useful therapeutic approach to treat this disease.56 According to the RM analysis, we infer that, regulation of inflammation may be the main role of JU in treating cerebral ischemia which also contributes to its synergistic mechanism from a convergent pattern.
Figure 8.
Pathway map of NOD-like receptor signaling in independent JU-RME. List genes are marked in red star. (A color version of this figure is available in the online journal.)
In summary, our drug RMs combining topology and function enable comprehension of the overlapping and diverse pharmacological mechanisms of different compounds individually and in combinations in treatment of cerebral ischemia. Specifically, we found that the additive effect of BJ may be resulted from multiple responsive modules working in a divergent pattern, and the synergistic effect of JU may be explained by a “central hit strategy”59 with fewer responsive modules regulating inflammation in a convergent pattern against cerebral ischemia. Nevertheless, limitations still exist. Firstly, we present only the emerged and lost modules to reflect drug therapy. Drug response module may also incorporate shrinkage, growth, split and other dynamic processes.60 Secondly, although the cerebral ischemic information carried by RMs has been validated by previous literatures, experimental validation should also be taken into account in future studies.
Supplementary Material
Acknowledgements
We thank the SysBiomics Bioinformatics Co. Ltd. for their help in performing the statistical and bioinformatics analyses. This study was supported by National 11th Five-year plan Supporting R&D Project (2006BAI08B04-06) and the 9th autonomously selected subject projects of the China Academy of Chinese Medical Sciences (Z0405).
Authors’ contributions
All authors participated in the design, interpretation of the studies and analysis of the data and review of the manuscript; as the guarantor, ZW conceived the study; YNY, ZW, and XXZ initially drafted the paper with subsequent contributions from all authors; BL, YYZ, JL, HXL, and YYC performed the research; PQW, RXK and HLW collected and cleaned the data.
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Cabusora L, Sutton E, Fulmer A, Forst CV. Differential network expression during drug and stress response. Bioinformatics 2005; 21: 2898–905. [DOI] [PubMed] [Google Scholar]
- 2.Wang X, Dalkic E, Wu M, Chan C. Gene module level analysis: identification to networks and dynamics. Curr Opin Biotechnol 2008; 19: 482–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao S, Iyengar R. Systems pharmacology: network analysis to identify multiscale mechanisms of drug action. Ann Rev Pharmacol Toxicol 2012; 52: 505–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang ZL, Wu WC, Liu JQ, Yao YB, Pan MD, Yang CB, Wang JG, Huang XW, Lin JY. Screening of differentially expressed genes related to ischemic stroke and functional analysis with DNA microarray. Eur Rev Med Pharmacol Sci 2014; 18: 1181–8. [PubMed] [Google Scholar]
- 5.Thieffry D, Romero D. The modularity of biological regulatory networks. Biosystems 1999; 50: 49–59. [DOI] [PubMed] [Google Scholar]
- 6.Wong DJ, Chang HY. Learning more from microarrays: insights from modules and networks. J Invest Dermatol 2005; 125: 175–82. [DOI] [PubMed] [Google Scholar]
- 7.Wagner GP, Pavlicev M, Cheverud JM. The road to modularity. Nat Rev Genet 2007; 8: 921–31. [DOI] [PubMed] [Google Scholar]
- 8.Girvan M, ME N. Community structure in social and biological networks. Proc Natl Acad Sci USA 2002; 99: 7821–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Z, Wang YY. Modular pharmacology: deciphering the interacting structural organization of the targeted networks. Drug Discov Today 2013; 18: 560–6. [DOI] [PubMed] [Google Scholar]
- 10.Wang Z, Liu J, Yu Y, Chen Y, Wang Y. Modular pharmacology: the next paradigm in drug discovery. Expert Opin Drug Discov 2012; 7: 667–77. [DOI] [PubMed] [Google Scholar]
- 11.Dimitrakopoulou K, Dimitrakopoulos GN, Sgarbas KN, Bezerianos A. Tamoxifen integromics and personalized medicine: dynamic modular transformations underpinning response to tamoxifen in breast cancer treatment. OMICS 2014; 18: 15–33. [DOI] [PubMed] [Google Scholar]
- 12.Zeng T, Wang DC, Wang X, Xu F, Chen L. Prediction of dynamical drug sensitivity and resistance by module network rewiring-analysis based on transcriptional profiling. Drug Resist Updat 2014; 17: 64–76. [DOI] [PubMed] [Google Scholar]
- 13.Iskar M1, Zeller G, Blattmann P, Campillos M, Kuhn M, Kaminska KH, Runz H, Gavin AC, Pepperkok R, van Noort V, Bork P. Characterization of drug-induced transcriptional modules: towards drug repositioning and functional understanding. Mol Syst Biol 2013; 9: 662–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sim SC, Ingelman-Sundberg M. Pharmacogenomic biomarkers: new tools in current and future drug therapy. Trends Pharmacol Sci 2011; 32: 72–81. [DOI] [PubMed] [Google Scholar]
- 15.Yu S, Zheng L, Li Y, Li C, Ma C, Yu Y, Li X, Hao P. Causal co-expression method with module analysis to screen drugs with specific target. Gene 2013; 518: 145–51. [DOI] [PubMed] [Google Scholar]
- 16.Lv L, Liu Y, Shi HF, Dong Q. Qingkailing injection attenuates apoptosis and neurologic deficits in a rat model of intracerebral hemorrhage. J Ethnopharmacol 2009; 125: 269–73. [DOI] [PubMed] [Google Scholar]
- 17.Zhou C, Wang Z, Jing Z, Zhang Z. Differences in pharmacological pathways among Qingkailing effective component. Chin Pharmacol Bull 2010; 4: 547–51. [Google Scholar]
- 18.Chen Y, Meng F, Fang H, Yu Y, Liu J, Jing Z, Lv A, Wang Z, Wang Y. Hierarchical profiles of signaling pathways and networks reveal two complementary pharmacological mechanisms. CNS Neurol Disord Drug Targets 2013; 12: 882–93. [DOI] [PubMed] [Google Scholar]
- 19.Chen Y, Zhou C, Yu Y, Liu J, Jing Z, Lv A, Meng F, Wang Z, Wang Y. Variations in target gene expression and pathway profiles in the mouse hippocampus following treatment with different effective compounds for ischemia-reperfusion injury. Naunyn Schmiedebergs Arch Pharmacol 2012; 385: 797–806. [DOI] [PubMed] [Google Scholar]
- 20.Liu J, Yin F, Zheng X, Jing J, Hu Y. Geniposide, a novel agonist for GLP-1 receptor.; prevents PC12 cells from oxidative damage via MAP kinase pathway. Neurochem Int 2007; 51: 361–69. [DOI] [PubMed] [Google Scholar]
- 21.Liu J, Zhou CX, Zhang ZJ, Wang LY, Jing ZW, Wang Z. Synergistic mechanism of gene expression and pathways between jasminoidin and ursodeoxycholic acid in treating focal cerebral ischemia-reperfusion injury. CNS Neurosci Ther 2012; 18: 674–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang YY, Li HX, Chen YY, Fang H, Yu YN, Liu J, Jing ZW, Wang Z, Wang YY. Convergent and divergent pathways decoding hierarchical additive mechanisms in treating cerebral ischemia-reperfusion injury. CNS Neurosci Ther 2014; 20: 253–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, Zhang ZJ, Zhou CX, Wang Y, Cheng YY, Darrel Duan DY, Wang YY, Wang Z. Outcome-dependent global similarity analysis of imbalanced core signaling pathways in ischemic mouse hippocampus. CNS Neurol Disord Drug Targets 2012; 11: 1070–82. [DOI] [PubMed] [Google Scholar]
- 24.Yellaboina S, Dudekula DB, Ko MSh. Prediction of evolutionarily conserved interologs in Musmusculus. BMC Genomics 2008; 9: 465–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aranda B, Achuthan P, Alam-Faruque Y, Armean I, Bridge A, Derow C, Feuermann M, Ghanbarian AT, Kerrien S, Khadake J, Kerssemakers J, Leroy C, Menden M, Michaut M, Montecchi-Palazzi L, Neuhauser SN, Orchard S, Perreau V, Roechert B, van Eijk K, Hermjakob H. The IntAct molecular interaction database in 2010. Nucleic Acids Res 2010; 38(Database issue): D525–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stark C, Breitkreutz BJ, Chatr-Aryamontri A, Boucher L, Oughtred R, Livstone MS, Nixon J, Van Auken K, Wang X, Shi X, Reguly T, Rust JM, Winter A, Dolinski K, Tyers M. The Bio GRID Interaction Database: 2011 update. Nucleic Acids Res 2011; 39(Database issue): D698–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ceol A, Chatr Aryamontri A, Licata L, Peluso D, Briganti L, Perfetto L, Castagnoli L, Cesareni G. MINT, the molecular interaction database: 2009 update. Nucleic Acids Res 2010; 38(Database issue): D532–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kohl M, Wiese S, Warscheid B. Cytoscape: software for visualization and analysis of biological networks. Methods Mol Biol 2011; 696: 291–303. [DOI] [PubMed] [Google Scholar]
- 29.Kenley EC, Cho YR. Entropy-based graph clustering: application to Biological and social networks. In: IEEE 11th international conference on data mining, 11–14 December 2011, Vancouver: IEEE, pp. 1116–1121.
- 30.Tan YJ, Wu J. Network structure entropy and its application to scale-free networks. Syst Eng Theor Pract 2004; 6: 001–001. [Google Scholar]
- 31.Li M, Chen JE, Wang JX. Research on robustness of PPI networks based on normalized entropy. Appl Res Comput 2009; 1: 029–029. [Google Scholar]
- 32.Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G. Go::termfinder-open source software for accessing gene ontology information and finding significantly enriched gene ontology terms associated with a list of genes. Bioinformatics 2004; 20: 3710–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flight RM, Wentzell PD. Potential bias in go:: termfinder. Brief Bioinform 2009; 10: 289–94. [DOI] [PubMed] [Google Scholar]
- 34.Gene Ontology Consortium. The gene ontology in 2010: extensions and refinements. Nucleic Acids Res 2010; 38(Database issue): D331–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blake JA, Bult CJ, Kadin JA, Richardson JE, Eppig JT. Mouse Genome Database Group. The Mouse Genome Database (MGD): premier model organism resource for mammalian genomics and genetics. Nucleic Acids Res 2011; 39(Database issue): D842–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4: 44–57. [DOI] [PubMed] [Google Scholar]
- 37.Wang Z, Jing ZW, Zhou CX, Zhang L, Cheng J, Zhang ZJ, Liu J, Xu CS, Li PT, Wang YY. Fusion of core pathways reveals a horizontal synergistic mechanism underlying combination therapy. Eur J Pharmacol 2011; 667: 278–86. [DOI] [PubMed] [Google Scholar]
- 38.Justicia C, Gabriel C, Planas AM. Activation of the JAK/STAT pathway following transient focal cerebral ischemia: signaling through Jak1 and Stat3 in astrocytes. Glia 2000; 30: 253–70. [DOI] [PubMed] [Google Scholar]
- 39.Chen YY, Yu YN, Zhang YY, Li B, Liu J, Li DF, Wu P, Wang J, Wang Z, Wang YY. Quantitative determination of flexible pharmacological mechanisms based on topological variation in mice anti-ischemic modular networks. PLoS One 2016;11:e0158379. [DOI] [PMC free article] [PubMed]
- 40.Kimberly WT, Wang Y, Pham L, Furie KL, Gerszten RE. Metabolite profiling identifies a branched chain amino acid signature in acute cardioembolic stroke. Stroke 2013; 44: 1389–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Delille HK, Bonekamp NA, Schrader M. Peroxisomes and disease-an overview. Int J Biomed Sci 2006; 2: 308–14. [PMC free article] [PubMed] [Google Scholar]
- 42.Wang A, Huang K, Shen Y, Xue Z, Cai C, Horvath S, Fan G. Functional modules distinguish human induced pluripotent stem cells from embryonic stem cells. Stem Cells Dev 2011; 20: 1937–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cha K, Kim MS, Oh K, Shin H, Yi GS. Drug similarity search based on combined signatures in gene expression profiles. Healthc Inform Res 2014; 20: 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yuasa T, Takahashi S, Hatake K, Yonese J, Fukui I. Biomarkers to predict response to sunitinib therapy and prognosis in metastatic renal cell cancer. Cancer Sci 2011; 102: 1949–57. [DOI] [PubMed] [Google Scholar]
- 45.Cohen AL, Soldi R, Zhang H, Gustafson AM, Wilcox R, Welm BE, Chang JT, Johnson E, Spira A, Jeffrey SS, Bild AH. A pharmacogenomic method for individualized prediction of drug sensitivity. Mol Syst Biol 2011; 7: 513–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lord CJ, Ashworth A. The DNA damage response and cancer therapy. Nature 2012; 481: 287–94. [DOI] [PubMed] [Google Scholar]
- 47.Luscombe NM, Babu MM, Yu H, Snyde M, Teichmann SA, Gerstein M. Genomic analysis of regulatory network dynamics reveals large topological changes. Nature 2004; 431: 308–12. [DOI] [PubMed] [Google Scholar]
- 48.Langfelder P, Luo R, Oldham MC, Horvath S. Is my network module preserved and reproducible? PLoS Comput Biol 2011; 7: e1001057–e1001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nizon M, Boutron A, Boddaert N, Slama A, Delpech H, Sardet C, Brassier A, Habarou F, Delahodde A, Correia I, Ottolenghi C, de Lonlay P. Leukoencephalopathy with cysts and hyperglycinemia may result from NFU1 deficiency. Mitochondrion 2014; 15: 59–64. [DOI] [PubMed] [Google Scholar]
- 50.Packer L, Tritschler HJ, Wessel K. Neuroprotection by the metabolic antioxidant alpha-lipoic acid. Free Radic Biol Med 1997; 22: 359–78. [DOI] [PubMed] [Google Scholar]
- 51.Connell BJ, Saleh M, Khan BV, Saleh TM. Lipoi acid protects against reperfusion injury in the early stages of cerebral ischemia. Brain Res 2011; 1375: 128–36. [DOI] [PubMed] [Google Scholar]
- 52.Kimberly WT, Wang Y, Pham L, Furie KL, Gerszten RE. Metabolite profiling identifies a branched chain amino acid signature in acute cardioembolic stroke. Stroke 2013; 44: 1389–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tomasevic G, Kamme F, Wieloch T. Changes in proliferating cell nuclear antigen, a protein involved in DNA repair, in vulnerable hippocampalneurons following global cerebral ischemia. Brain Res Mol Brain Res 1998; 60: 168–76. [DOI] [PubMed] [Google Scholar]
- 54.Vries B, Walter SJ, Wolfs TG, Hochepied T, Räbinä J, Heeringa P, Parkkinen J, Libert C, Buurman WA. Exogenous alpha-1-acid glycoprotein protects against renal ischemia-reperfusion injury by inhibition of inflammation and apoptosis. Transplantation 2004; 78: 1116–24. [DOI] [PubMed] [Google Scholar]
- 55.Downes CE, Wong CH, Henley KJ, Guio-Aguilar PL, Zhang M, Ates R, Mansell A, Kile BT, Crack PJ. MyD88 is a critical regulator of hematopoietic cell-mediated neuroprotection seen after stroke. PLoS One 2013; 8: e57948–e57948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu H, Wei X, Kong L, Liu X, Cheng L, Yan S, Zhang X, Chen L. NOD2 is involved in the inflammatory response after cerebral ischemia-reperfusion injury and triggers NADPH oxidase 2-derived reactive oxygen species. Int J Biol Sci 2015; 11: 525–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Muir KW, Tyrrell P, Sattar N, Warburton E. Inflammation and ischaemic stroke. Curr Opin Neurol 2007; 20: 334–42. [DOI] [PubMed] [Google Scholar]
- 58.Amantea D, Nappi G, Bernardi G, Bagetta G, Corasaniti MT. Post-ischemic brain damage: pathophysiology and role of inflammatory mediators. FEBS J 2009; 276: 13–26. [DOI] [PubMed] [Google Scholar]
- 59.Liu J, Wang Z. Diverse array-designed modes of combination therapies in Fangjiomics. Acta Pharmacol Sin 2015; 36: 680–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Palla G, Barabási AL, Vicsek T. Quantifying social group evolution. Nature 2007; 446: 664–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.