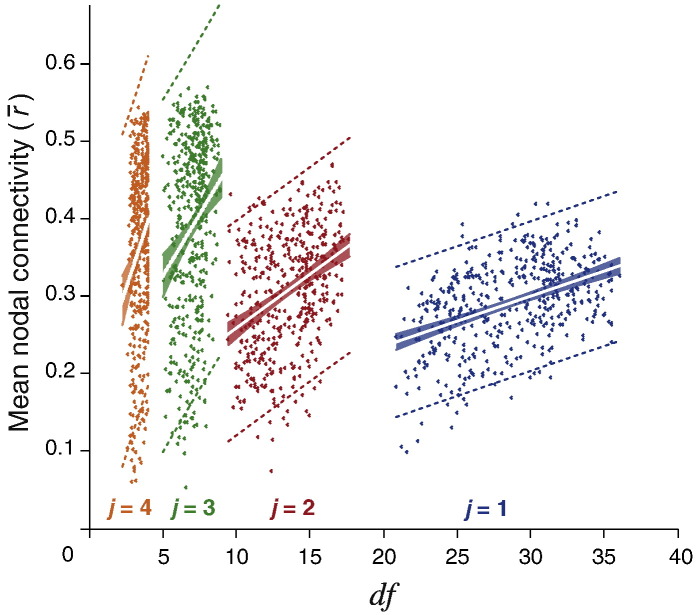
Fig. 3.
Mean nodal connectivity shows strong linear dependence on the df. Voxel-wise time series were parcellated into 470 regions and a connectivity matrix was generated for each wavelet scale (frequency band) by computing the Pearson correlation r on MODWT wavelet coefficients, between each pairwise combination of nodes. The mean nodal connectivity represents the average of all non-zero correlations between a given node and all other nodes in the brain. The voxel-wise df estimate was generated for each wavelet scale, by wavelet despiking. The nodal df were computed by averaging the df of all voxels in each region (defined by the 470 region parcellation template), at each scale. This resulted in an df estimate for each of 470 regions at different wavelet scales. The mean nodal connectivity () was then plotted against the nodal df for the highest four wavelet scales (j), representing the most commonly analyzed frequency bands: j = 1, 0.13 < f < 0.25 Hz; j = 2, 0.06 < f < 0.13 Hz; j = 3, 0.03 < f < 0.06 Hz; j = 4, 0.02 < f < 0.03 Hz. The mean nodal connectivity showed strong linear dependence on the df at all four wavelet scales (P < 0.0001, F test).