Table 1.
Host-guest binding free energies. The OSRW column presents the average of results from the full length of simulations, while the OSRW (10 ns) column presents values cut off at 10ns. The host molecule for all structures is cucurbit[7]uril. All free energies are given in kcal/mol. The experimental free energies hold an uncertainty of ± 0.1 kcal/mol.
| Guest | Guest Structure |
Δ
G
bind
|
|||
|---|---|---|---|---|---|
| Experimental | BAR | OSRW | OSRW (10 ns) | ||
| C1 |

|
−9.9 | −12.27 ± 0.92 | −11.64 ± 0.42 | −10.43 ± 1.01 |
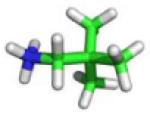
| C2 |
|
−9.6 | −6.46 ± 0.65 | −7.29 ± 0.20 | −5.50 ± 0.05 |
| C3 |

|
−6.6 | −6.59 ± 0.74 | −6.71 ± 0.74 | −5.17 ± 1.03 |
| C4 |

|
−8.4 | −11.34 ± 0.89 | −10.02 ± 0.25 | −8.31 ± 0.75 |
| C5b |

|
−8.5 | −3.39 ± 0.97 | −5.00 ± 0.11 | −4.46 ± 0.26 |
| C6 |

|
−7.9 | −6.18 ± 0.69 | −7.01 ± 0.35 | −6.64 ± 0.07 |
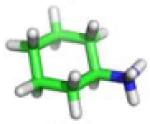
| C7 |
|
−10.1 | −10.49 ± 0.66 | −10.05 ± 0.09 | −10.96 ± 0.78 |
| C8 |

|
−11.8 | −11.84 ± 0.68 | −11.44 ± 0.09 | −11.15 ± 0.76 |
| C9 |

|
−12.6 | −15.42 ± 0.71 | −15.35 ± 0.29 | −15.44 ± 0.65 |
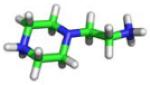
| C10 |
|
−7.9 | −5.06 ± 0.91 | −3.90 ± 0.26 | −3.69 ± 0.68 |
| C11 |

|
−11.1 | −10.48 ± 0.64 | −10.06 ± 0.25 | −9.82 ± 0.34 |
| C12 |

|
−13.3 | −12.11 ± 0.70 | −12.57 ± 0.03 | −12.33 ± 0.16 |
| C13 |

|
−14.1 | −13.92 ± 0.65 | −13.13 ± 0.13 | −12.63 ± 0.52 |
| C14 |

|
−11.6 | −12.41 ± 0.72 | −12.75 ± 0.59 | −12.05 ± 0.58 |
