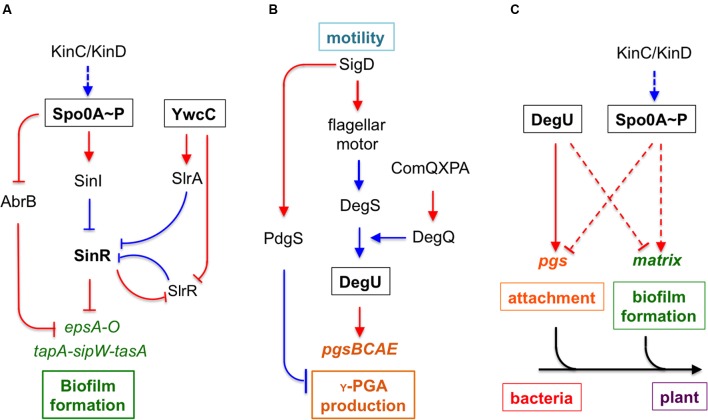
FIGURE 1.
An integrated model for regulation and function of poly-γ-glutamic acid (γ-PGA) and biofilm formation during Bacillus subtilis–plant interactions. (A) A schematic drawing of the regulatory pathways in B. subtilis that control biofilm matrix genes. KinC and KinD are two sensory histidine kinases involved in sensing environmental signals. KinC and KinD activate the master regulator Spo0A through protein phosphorylation. Phosphorylated Spo0A in turn activates biofilm matrix genes via two independent mechanisms, one through the transition stage regulator AbrB and the other through the SinI-SinR regulatory module. SinR is a biofilm master repressor of the matrix genes. YwcC is a TetR-type repressor, which controls genes encoding two antagonist proteins (SlrA and SlrR) for SinR. The epsA-O and the tapA-sipW-tasA operons are involved in making the EPS and the TasA amyloid fibers, respectively, of the biofilm matrix. (B) The pgsBCAE operon for γ-PGA biosynthesis is chiefly regulated by the two-component system DegS-DegU. Another two-component system ComA-ComP regulates the degQ gene, which encodes a small regulator for DegU activation. DegU is also activated by a yet unknown mechanism in sensing flagellar motion. (C) The interplay between the control on biofilm formation and that of γ-PGA production. Response regulators DegU and Spo0A both mediate opposing regulation on γ-PGA biosynthesis genes and genes for biofilm matrix production in two independent pathways. Dashed lines indicate indirect regulations. We also propose that γ-PGA and biofilm matrix may play sequential roles in the attachment of the bacteria to the root surface and in formation of root-surface associated biofilms during B. subtilis–plant interactions. Lines in red indicate gene regulation while lines in blue indicate protein–protein interactions.