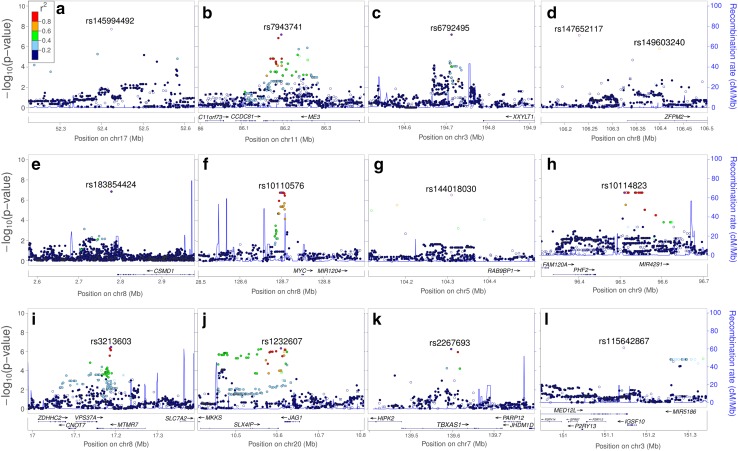
Fig. 2.
Locus-specific plots highlighting the loci implicated by SNPs reaching a significance threshold of meta P value <10−6 for FA. a–i Each plot shows the −log10 P value (Y-axis) of SNPs arranged according to their chromosomal positions (X-axis). The locus-specific plots include the genes ME3 (b), ZFPM2 (d), MTMR7 (i), JAG1 and SLX4IP(j), TBXAS1 (k), and IGSF10 and MED12L (l). The blue lines show estimated recombination rates calculated from the HapMap data. The arrows represent the genomic locations of genes based on the NCBI Build 37 human assembly. SNP color represents LD with the SNP showing highest association in the locus. The SNP annotation is represented as follows: circles no annotation; squares synonymous or 3′ UTR; triangles non-synonymous; asterisks TFBScons (in a conserved region predicted to be a transcription factor binding site); squares with an X, MCS44 placental (in a region highly conserved in placental mammals)