Abstract
The conserved coat or V2 gene of begomoviruses is responsible for viral movement in the plant cells. RNAi technology was used to silence V2 gene for resistance against these viruses in transgenic plants. The transformation of the RNAi-based gene construct targeting V2 gene of CLCuKoV-Bur, cloned under 35S promoter, was done in two elite cotton varieties MNH-786 and VH-289 using shoot apex cut method of gene transformation. The transformation efficiency was found to be 3.75 and 2.88 % in MNH-786 and VH-289, respectively. Confirmation of successful transformation was done through PCR in T 0, T 1, and T 2 generations using gene-specific primers. Transgenic cotton plants were categorized on the basis of the virus disease index in T 1 generation. Copy number and transgene location were observed using FISH and karyotyping in T 2 generation which confirmed random integration of V2 RNAi amplicon at chromosome 6 and 16. Real-time quantitative PCR analyses of promising transgenic lines showed low virus titer compared to wild-type control plants upon challenging them with viruliferous whiteflies in a contained environment. From the results, it was concluded that amplicon V2 RNAi construct was able to limit virus replication and can be used to control CLCuV in the field.
Keywords: Gene transformation, siRNA, CLCuD, Begomovirus, V2 ORF, Knockdown
Introduction
The agriculture sector is of utmost importance for Pakistan’s economy as it contributes 21.4 % of the overall GDP. Among major crops, only cotton shares 1.2 % of the country’s overall GDP and contributes more than 60 % of its foreign exchange. Cotton leaf curl disease (CLCuD) is a major threat to the production of cotton in Pakistan [1–3]. CLCuD remained a sporadic nuisance before 1986, but in the subsequent years, it rapidly spread across the cotton growing areas of Pakistan and became an epidemic in 1991–2 causing heavy yield losses during those years. During the late 1990s, resistant cotton varieties were introduced and thus losses due to this disease reduced [4]. However, in the year 2001–2, in the Burewala region of Punjab province, resistant cotton varieties began to show typical symptoms of CLCuD. This was an indication of a second epidemic and referred to as Cotton leaf curl Burewala virus which is now known as Cotton leaf curl Kokhran virus-Burewala strain (CLCuKoV-Bur). This virus has spread and infection is now found in most cotton growing areas in Pakistan [5, 6].
Typical symptoms of CLCuD include thickening, darkening and swelling of veins, upward or downward curling of leaves, and enations (cup-shaped laminar outgrowths on the undersides of leaves) [7–9]. The CLCuD is caused by a complex consisting of several monopartite begomoviruses (family Geminiviridae) that essentially require a satellite molecule known as cotton leaf curl Multan beta satellite (CLCuMB). CLCuMB is entirely dependent on the helper begomovirus for its replication and encapsulation, while it acts as a pathogenicity determinant and encodes protein βC1 that can overcome host defense responses [10]. It has been shown that CLCuKoV-Bur is a recombinant molecule derived from two previously reported viruses i.e., Cotton leaf curl Multan virus and Cotton leaf curl Kokhran virus [6]. Similarly, the beta satellite associated with CLCuKoV is a recombinant of CLCuMB and Tomato leaf curl beta satellite [11]. Also associated with the disease is another self-replicating component referred to as an alpha satellite. The role of the alpha satellite is not fully known; however, the rep protein of an alpha satellite has been shown to be a suppressor of gene silencing [6]. These viral components are transmitted to the plant by whitefly (Bemisia tabaci) [12].
To date, no natural resistance to immunity is available in cotton against CLCuD. Besides conventional methods, the use of biotechnological approaches can be a possible solution to this problem, based on RNA interference (RNAi) in varieties with an agronomical CLCuD-tolerant background [13–15]. RNAi (or gene silencing) is a homology based down-regulation/silencing of genes mechanism which is evolutionary conserved and works in a sequence-specific manner. Begomoviruses can be targeted by gene silencing both at the transcriptional level [transcriptional gene silencing (TGS)], which results in methylation of viral DNA, and the post-transcriptional level [post-transcriptional gene silencing (PTGS)], which results in degradation of viral transcripts. RNAi is always triggered by a dsRNA that is cleaved into short interfering siRNAs by an RNase referred to as DICER-like. The siRNAs then guide sequence-specific silencing. For PTGS, siRNAs are incorporated into an enzyme complex, the RNA-induced silencing complex which degrades homologous mRNAs [16–19]. Resistance against viruses in plants can be obtained by inducing RNAi in plants through the introduction of a sequence homologous to the virus in the form of a hairpin (hp) [20]. One of the advantages of RNAi is the silencing signal which is not limited to individual cells, but can spread to neighboring cells and more distant tissues [21, 22].
RNA interference is an emerging technology for developing insect resistance genes. Many transgenic RNAi-based genetically modified (GM) plants, targeting insects have been developed, including GM corn, GM rice, and GM cotton, and additional GM crops are in the process of development. Various researchers have tried to use RNAi technology to obtain resistance against geminiviruses [23–26]. Recently, RNAi-based resistance has been successfully applied in beans against Bean golden mosaic virus in Brazil [27, 28].
In this study, two elite cotton varieties i.e., MNH-786 and VH-289 were transformed with an amplicon RNAi construct against V2 gene of CLCuKoV-Bur. The philosophy of silencing of this gene was to restrict virus movement and further spread. Similar efforts were made for transforming Indian cotton varieties with RNAi gene constructs targeting V2 and Intergenic region (IR) [15, 29].
Materials and Methods
Gene Construct
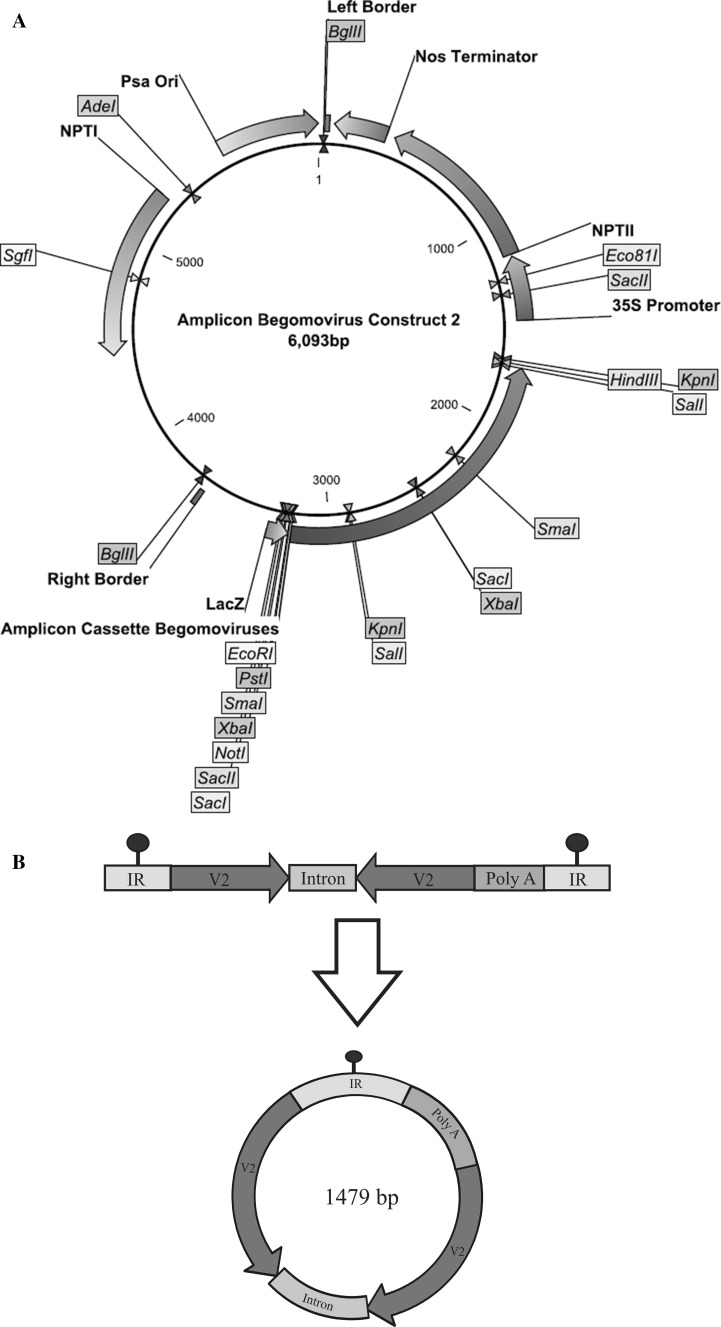
Two hundred and forty Nucleotides from V2 of Cotton leaf curl Kokhran virus-Burewala (CLCuKoV-Bur; Accession No. AM421522) were taken to make hairpin construct synthetically. Both sense and antisense sequences of V2 are separated by 115 nucleotides from an intron of Mungbean yellow mosaic India virus (MYMIV), Accession No. FM202439, to induce hairpin. The IR of CLCuMuV (Accession No. AY312430) was used, which presumably contains both the Rep promoter and the viral origin of replication. The IR, which is used for higher expression of siRNAs, consists of 287 nt [30]. The only way this amplicon construct can work is by having two IRs for replicational release by the Rep of the infecting virus. However, to make the construct as a defective interfering molecule, 247 nt from Poly A of Cotton leaf curl Multan alpha satellite (CLCuMuA), accession no. AJ132344, were added after the hairpin construct. As a result, the total size of the construct would be 1.479 kb which is approximately equal to the defective molecules of DNA A of begomoviruses.
The construct contains two IRs that enable the construct to be replicationally released from the plant genome, circularized, and replicated by the Rep of the infecting begomovirus. Thus, the construct will constitutively express siRNA derived from V2 gene and will replicate as an episome upon virus infection. The construct was initially cloned in pTZ57R/T vector and subsequently; the fragment was directionally cloned at Hind III and EcoRI sites into the binary vector pGreen0029. This construct will be called as amplicon V2 RNAi in the manuscript (Fig. 1a, b).
Fig. 1.
Vector carrying Amplicon-Based RNAi gene targeting Begomoviruses
Primer Designing
Internal primers were designed for the construct (primer details are given in Table 1).
Table 1.
Internal primer details for gene amplification of RNAi gene construct and real-time PCR
| Name | Primer sequence | Length | Tm | Product size (bp) |
|---|---|---|---|---|
| C2-int (L) | TCATAATCTAAACCAAACAGGGAAA | 25 | 60.11 | 540 |
| C2-int (R) | TTACAATCAGGTCCTTCAGCAAA | 23 | 60.99 | |
| ClCuV Burewala (F) | CGAAAGAAGAAGGAGAAAAA | 23 | 53.0 | 203 |
| ClCuV Burewala (R) | AGCAAGAGGAGGACAGCAGA | 20 | 59.4 | |
| ClCuV Multan beta (F) | GTTCCGCTGGTTGTCATTTC | 20 | 55.4 | 268 |
| ClCuV Multan beta (R) | CCTCTTCAGTTCCGTTTTTC | 21 | 54.6 |
Bold values are the important facts and figures about results
Confirmation of Amplicon-Based Begomovirus Construct
The amplicon V2 RNAi construct was confirmed through amplification using gene-specific primers. The construct was also confirmed, using plasmid as template, through restriction digestion analysis. Furthermore, confirmation of successful electroporation in Agrobacterium was also done through colony PCR using gene-specific primers.
Plant Material
On request, seeds of cotton varieties MNH 786 and VH 289 were provided by the Cotton Research Institute, Multan and Cotton Research Institute, Vehari, respectively.
Plant Transformation
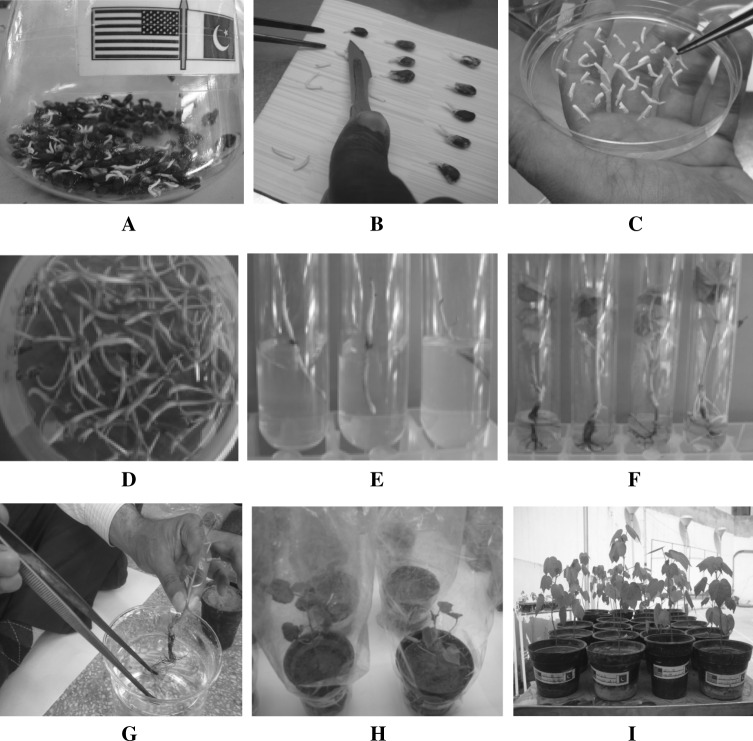
Amplicon V2 RNAi was transformed in embryos via Agrobacterium transformation through the Embryo shoot apex cut method as described by Rao et al. [31, 32]. A total of 14200 embryos were used in transformation experiments from which 4000 were of control. The plantlets were given kanamycin selection (100 µg/ml) in the shoot development medium (Murashige and Skoog medium MS: 4.43 g/L; Sucrose: 30 g/L; Kinetin: 50 mg/L; Phytagel 3 g/L; pH: 5.8). While in the root development, media was also supplemented with the growth hormones IAA (1 mg/L) and IBA (1 mg/L). The plants were shifted to pots after proper development of shoots and roots. The soil mixture used in pots was of the same composition as described by Rao et al. [32]. When plants were able to tolerate 6 h sunlight, they were shifted to the field (Figs. 2 and 3). The seeds of T 0 PCR-positive transgenic plants were used to rise T 1 generation, while the seeds of confirmed transgenic cotton plants of T 1 generation were then used to advance T 2 generation.
Fig. 2.
Steps from embryo transformation to shifting of plants in soil pots a Germination of Embryos in sterilized flasks, b Embryos isolation from seeds, c Agrobacterium-treated embryos are co-cultivated on MS medium, d Agrobacterium-treated embryos growing on MS medium (4 days), e Inoculation of embryos in test tubes containing MS media with kanamycin selection, f Roots developing in MS rooting medium, g Transgenic plants being shifted in soil pots, h putative transgenic plants in soil pots covered with polythene bags, i Acclimatization of putative transgenic plants
Fig. 3.
Putative (T 0) plants in field a Newly shifted T 0 transgenic plants, b T 0 plants thriving in the field, c selfing of bolls to self-cross the seeds, d T 0 bolls at maturity, e and f Picking of T 0 bolls
Molecular Analysis
Polymerase Chain Reaction (PCR)
The CTAB method was used for genomic DNA isolation of putative transgenic cotton plants [33]. Amplification of the transgene with specific primers was done by PCR to confirm the successful transformation. DNA isolated from putative plants was used as a template; the plasmid construct was used as positive control, while the DNA isolated from untransformed plants was used as negative control. The PCR master mix was composed of 3 µl (10 ng) of template DNA, 2 µl PCR Buffer (10×), 2.5 µl MgCl2, 2 µl forward primer (10 pmol), 2 µl reverse primer (10 pmol), 2µl dNTPs (2 mM), and 1 µl (1.25 units) of Taq DNA polymerase (Fermantas cat # EP0402). The PCR reaction was initiated with denaturation at 95 °C for 5 min and subjected to 35 cycles as follows: 95 °C for 1 min, 59 °C for 1 min, and 72 °C for 1 min. Extension phase was prolonged for 10 min at 72 °C. The transgenic plants were screened at T 0, T 1 and T 2 generation on the basis of PCR results.
Monitoring of Disease Symptoms and Determination of Viral Disease Index
Monitoring of CLCuV symptoms of transgenic cotton plants was done through random selection. None of the plant was sprayed with whitefly control. The disease index was calculated using a scale described by Akhtar and Khan [34]. The inoculation of transgenic along with control cotton plant was done by incubating each transgenic and control plant with ten viruliferous whiteflies (produced by feeding on symptomatic non-transgenic plants). PCR reaction was performed to confirm and select the infected whiteflies in controlled greenhouse condition, and observation were taken after 3 week-interval of plant inoculation with whitefly.
Percentage disease index was established by following the procedure determined by Farooq et al. [35] and applied in cotton by Akhtar et al. [36]. According to this formula, disease index of cotton line with a factor 100/6 in which 100 determined percentage and 6 determined total level of disease index [35, 36].
Virus Titer Determination Through Real-Time Quantitative PCR
The virus titer in different transgenic plants with amplicon V2 RNAi was determined through Thermo Scientific Maxima SYBR Green qPCR kit (cat# K0241). The DNA of transgenic and non-transformed plants was diluted 10× before using as a template. CLCuKoV-Bur plasmid construct and CLCuMB plasmid construct were used as the standard for absolute quantification, while virus-infected plants were used as positive control. The master mix contained 10 µl SYBER green, 0.35 µl of Forward primer (10 pmol, Table 1), 0.35 µl of Reverse primer (10 pmol, Table 1), and 8.3 µl of template DNA (100/reaction). The qPCR reaction was started with an initial denaturation at 95 °C for 10 min, 40 cycles of 95 °C for 15 s, 59 °C for 30 s, and 72 °C for 30 s. A final extension was given at 72 °C for 10 min.
Southern Blot Analysis
The transgene copy no was determined using the Southern blot analysis as described by Southern [37]. Genomic DNA from apical leaves of putative transgenic cotton plants and untransformed control plants was isolated using Thermo scientific Genomic DNA purification kit (cat # K0512) by following the manufacturer’s guidelines. Genomic DNA (20 μg) was digested with HindIII enzyme according to the supplier’s instructions (Enzyme Production Lab of the National Centre of excellence in molecular biology (CEMB), Pakistan). The color was detected by 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium (BCIP/NBT) tablets (Sigma B5655) dissolved in water according to the manufacturer’s instruction.
Fluorescence In Situ Hybridization (FISH)
Labeling of the probe for transgene detection was done by Fluorescein ULS® Labeling Kit (Fermentas K0641) by following the instructions given by the manufacturer, and in situ hybridization was carried out according to protocol described by Rahman et al. [38] on metaphase chromosomal spreads.
Results
Transformation of RNAi Gene Construct in Cotton Embryos
The Agrobacterium shoot apex method of transformation optimized at CEMB was used to transform MNH-786 and VH-28 varieties of cotton. Eighty-six putative transgenic plants were obtained from 43 transgenic experiments of MNH-786, while 43 putative transgenic plants were obtained from 41 transgenic experiments of VH-289 (Table 2).
Table 2.
The record of transformation experiments done for RNAi construct transformation
| Variety | Experiments | Total seeds | No. of embryos isolated | No. of shoots in test tubes after 30 days | Acclimatization | Plants acclimatized and thrived in field | |
|---|---|---|---|---|---|---|---|
| No. of plants shifted to soil pots | No. of plants shifted to Field | ||||||
| MNH786 | 43 | 27,300 | 7400 | 278 | 86 | 24 | 12 |
| VH289 | 41 | 26,700 | 6800 | 196 | 43 | 26 | 11 |
Bold values are the important facts and figures about results
The high mortality rates from embryo inoculation till the development of rooted plantlets on kanamycin selection media showed low transformation efficiency rates i.e., 3.75 % in the case of MNH-786 and 2.88 % in case of VH-289 (Table 3).
Table 3.
Transformation Efficiency of Agrobacterium-mediated transformation
| Variety | No. of embryos isolated | No. of plants obtained after 8 weeks | Transformation efficiency (%) |
|---|---|---|---|
| MNH786 | 7400 | 278 | 3.75 |
| VH289 | 6800 | 196 | 2.88 |
Bold values are the important facts and figures about results
Confirmation of Gene Integration in T0, T1, and T2 Transgenic Plants
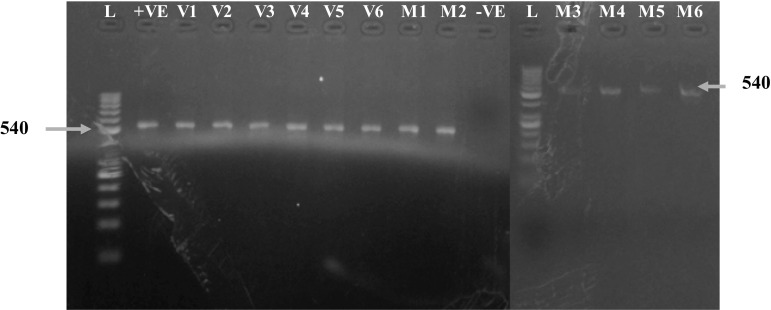
PCR analysis was performed to confirm the integration of the gene construct in three generations of transgenic plants. Eighty-six putative transgenic cotton plants of MNH-786 and 43 putative transgenic cotton plants of VH-289 were analyzed. Confirmation of successful transformation of putative transgenic cotton plants was done by PCR using plasmid as positive control and wild-type cotton plants as negative controls. PCR analysis (Fig. 4) confirmed the successful transformation of the amplicon V2 RNAi construct in these newly transformed cotton lines. The amplification of 540 bp internal fragments of RNAi construct in 26 putative transgenic plants of MNH-786 and 26 putative transgenic plants of VH-289 was achieved. No amplification was detected in negative control.
Fig. 4.
PCR analysis of putative transgenic plants (VH-289 & MNH-786) L 50 bp DNA ladder; −ve the non-transgenic plant, +ve plasmid construct was used as positive control, V1–V6 putative transgenic plants of VH-289, M1–M6 putative transgenic plants of MNH-786
A total of 11 MNH-786 and 12 VH-289 transgenic events were used for cultivation of the T 1 generation. The plants were also analyzed. PCR to amplify a 540 bp region of amplicon V2 RNAi construct was performed for T 1 transgenic cotton using plasmid as positive and wild-type cotton plants as negative controls with the same primers as were used for analysis of putative transgenic plants in T 0 generation. Seeds of transgenic T 1 events were used to rise the T 2 generation. The T 2 generation plants were tested through PCR. PCR analysis confirmed the presence of the amplicon V2 RNAi construct.
Viral Disease Index
The transgenic plants in the T 1 generation were found to be affected by the CLCuD infection on August 2013. Continuous observations were made for four consecutive months, starting from onset of disease. Monthly pictorial view of T 1 transgenic plants is shown in Fig. 5.
Fig. 5.
Pictorial View of Onset of CLCuV on T1 generation observed in different months. a Mild Symptoms of CLCuV on transgenic Cotton Plants. b Persistence of Mild Symptoms (CLCuV Tolerance) by Transgenic Cotton Plants. c Recovery of Plants vegetatively and reproductively while tolerating and minimizing viral titer. d Cotton Plants at Maturity showing tolerance of CLCuV with mild symptoms
To calculate the viral disease index, each T 1 transgenic plant was rated as per criteria described by Akhtar and Khan [34] (Table 4).
Table 4.
Viral disease Index of transgenic plants
| Symptoms | Disease ratings | Disease index (%) | Disease reaction |
|---|---|---|---|
| Absence of symptoms | 0 | 0 | Immune |
| Thickening of a few small veins or the presence of leaf enations on 10 or fewer leaves of plants | 1 | 0.1–1 | Highly resistant |
| Thickening of the small group of veins | 2 | 1.1–5 | Resistant |
| Thickening of veins, but no leaf curling | 3 | 5.1–10 | Moderately resistant |
| Severe vein thickening or leaf curling at the top of the third plant | 4 | 10.1–15 | Moderately susceptible |
| Severe vein thickening or leaf curling on half of the plant | 5 | 15.1–20 | Susceptible |
| Severe vein thickening, leaf curling, stunted growth of the plant and less fruit production | 6 | >20 | Highly susceptible |
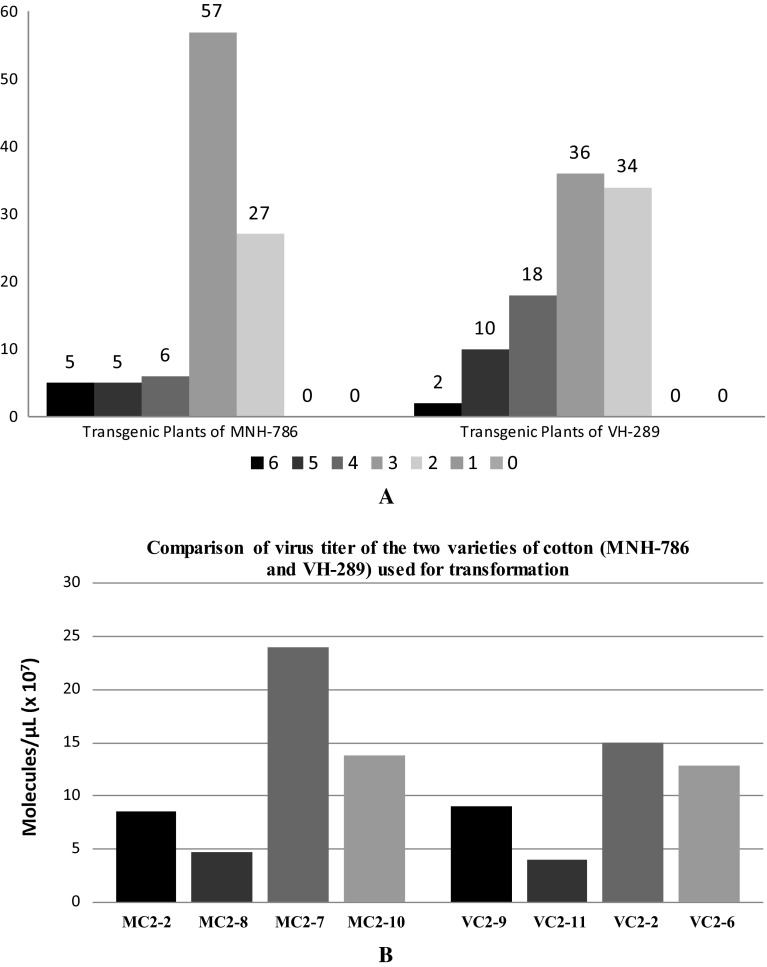
Different plants showed different ratings. The plants with maximum and minimum disease ratings were selected for viral titer determination through real-time quantitative PCR (qPCR). From ten transgenic VH-289 and MNH-786 lines along with non transgenic control lines, total one hundrad plants were screened. The number of plants of each variety with their disease ratings is given in Table 5.
Table 5.
Disease ratings of transgenic and non-transgenic plants
| Sr. No | Plants types | No. of plants under disease rating 6 | No. of plants under disease rating 5 | No. of plants under disease rating 4 | No. of plants under disease rating 3 | No. of plants under disease rating 2 | No. of plants under disease rating 1 | No. of plants under disease rating 0 |
|---|---|---|---|---|---|---|---|---|
| 1 | Non-transgenic control plants | 30 | 21 | 19 | 30 | 0 | 0 | 0 |
| 2 | Transgenic plants MNH-786 | 5 | 5 | 6 | 57 | 27 | 0 | 0 |
| 3 | Transgenic plants MVH-289 | 2 | 10 | 18 | 36 | 34 | 0 | 0 |
Virus Titer of T1 Transgenic Plants
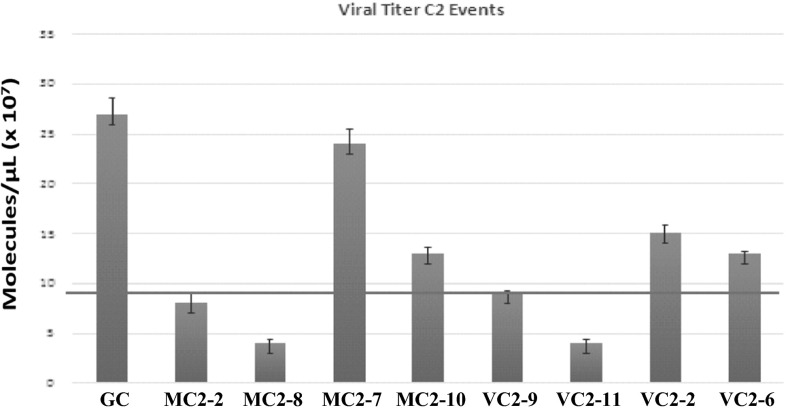
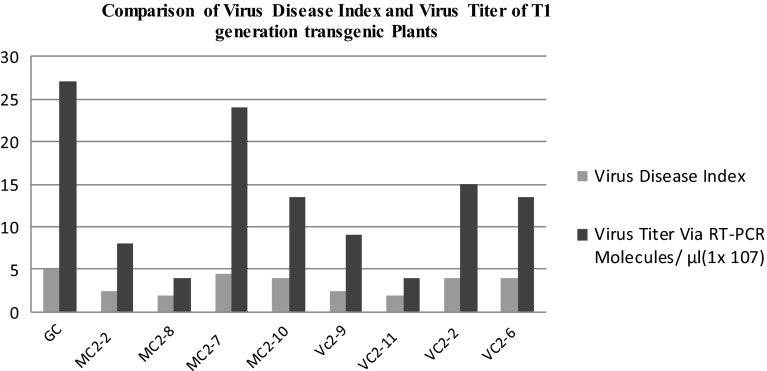
Absolute quantification via qPCR was used to determine the virus titer in eight different events of T 1 generation. A wild-type plant was used as negative control. It is obvious from Fig. 6 that all selected events showed a different virus titer. A tentative demarcation line was inserted (shown in green) to select the plants with minimum viral titer. The results were quite promising as the plants with lower disease index rating, i.e., MC2-2, MC2-8, VC2-9, and VC2-11 also showed low virus titer in qPCR.
Fig. 6.
Viral Titer calculation via real-Time PCR in T1 transgenic plants GC2 −ve control, MC2-2, MC2-8, MC2-7, and MC2-10 Transgenic plants of MNH-786; VC2-9, VC2-11, VC2-2, and VC2-6 transgenic plants of VH-289 in T1 generation harboring RNAi gene construct
The results of the CLCuD disease index and viral titer determined via qPCR were compared as shown in Fig. 7. It showed that symptom severity was directly proportional to virus titer. Plants having high virus titer exhibited more severe symptoms than those plants having low virus titer.
Fig. 7.
Comparison of viral disease index and virus titer of T1 generation transgenic Plants. GC non-transgenic plant, MC2-2, MC2-8, MC2-7, and MC2-10 Transgenic plants of MNH-786, VC2-9, VC2-11, VC2-2, and VC2-6 transgenic plants of VH-289
A comparative study of virus titer and virus disease index of both transgenic varieties was also performed (Fig. 8a, b).
Fig. 8.
a Comparison of disease index of the two varieties of cotton (MNH-786 and VH-289) used for transformation. Y-axis is showing number of plants, while X-axis is showing different disease ratings (0–6) as per Table 4. b Comparison of virus titer of the two varieties of cotton (MNH-786 and VH-289) used for transformation. Y-axis is showing virus titer in terms of molecules/µl × 107, while X-axis is showing transgenic plants, MC2-2, MC2-8, MC2-7, and MC2-10 of MNH-786 and VC2-9, VC2-11, VC2-2, and VC2-6 of VH-289
Location of Transgene in Cotton
Southern Blot Analysis
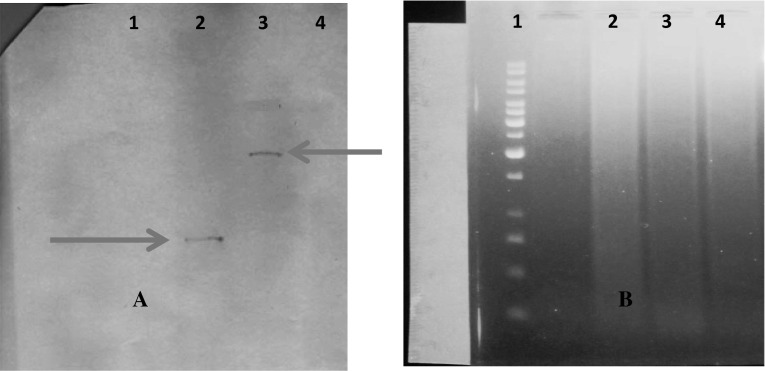
The stable integration of RNAi in the cotton plant genome and transgene copy number was confirmed by the Southern blot analysis. The copy number of RNAi transgene was obtained by a specific probe, which highlighted a different copy number based on restriction digestion of genomic DNA with unique sites using HindIII enzyme. The results clearly depict one copy number in transgenic cotton plants VC2-11 and MC2-8B respectively (Fig. 9).
Fig. 9.
a and b Southern Blot Analysis and Restriction digestion gel of Southern for RNAi in transgenic Cotton plant Lane 1 1 Kb DNA Ladder, Lane 2 MC2-8B transgenic cotton plant of MNH 786 variety, Lane 3 VC2-11 transgenic cotton plant of variety VH-289, Lane 4 Non-transgenic cotton control plant
Fluorescence In Situ Hybridization
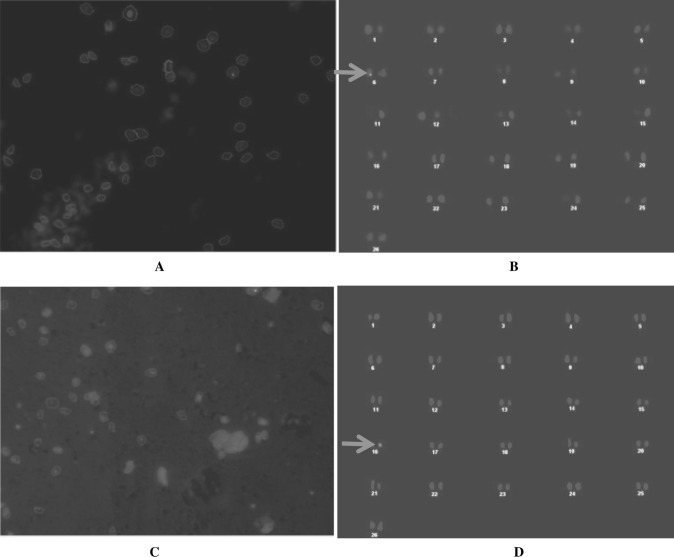
Integration of amplicon V2 RNAi in cotton plants was also confirmed by fluorescent in situ hybridization. There was random integration of the RNAi gene in Chromosome 6 and 16. One copy at Chromosome 6 was observed in transgenic event MC2-8 from the MNH 796 variety, while a single copy at chromosome 16 was observed in transgenic event VC2-11 from variety VH-289 (Fig. 10a–d).
Fig. 10.
a, b, c, and d Fluorescence in situ hybridization (FISH) of RNAi of two different plants Construct 2 i.e., MC2-8B (a and b) and VC2-11 (c and d). a Metastatic data of MC2-8B transgenic plant, b Karyotping of RNAi transgene of transgenic plant MC2-8B transgenic plant, c Metastatic data of VC2-11 transgenic plant, d Karyotping of RNAi transgene of transgenic plant VC2-11 transgenic plant
Discussion
Cotton is the white gold of Pakistan’s economy. Pakistan is the largest exporter of cotton yarn. Therefore the efforts to improve crop production, improve fiber quality, and pest management by genetic modification are critical. Cultivation of resistant crops and use of insecticides are currently the two major controls used for CLCuD [39]. According to Mansoor et al. [5], CLCuD-resistant crops were successfully developed in 1990s, but the virus soon developed resistance to these varieties [40]. An alternative strategy to control CLCuD could be RNAi. The results presented here have shown that the amplicon V2 RNAi approach, which is targeted against highly conserved V2 gene of the begomovirus, has the potential to provide resistance against CLCuKoV-Bur CLCuD in transgenic cotton plants. The majority of the transgenic cotton plants, upon challenge with viruliferous whiteflies in a contained environment, showed lower disease symptoms as well as disease index ratings. qPCR analyses showed the presence of virus in all transgenic plants, but there was a significant difference in virus titer compared with wild-type cotton plants. The level of resistance obtained here may be more accurately stated as highly tolerant to infection. These results are consistent with several studies that have exploited the RNAi approach for transgenic resistance against geminiviruses. According to Ali et al. [43], effective resistance in plants against monopartite begomoviruses can be obtained by applying a miRNA approach. [15, 41–44].
In the present study, the amplicon V2 RNAi gene construct was transformed in elite cotton varieties MNH-786 and VH-289. V2 protein in monopartite virus is a symptom determinant and elicited cell death. When expressed in plants using Potato virus X-based vector, it can act as a suppressor of gene silencing [45, 46]. Moreover, V2 encoded by CLCuMV is a very strong suppressor of gene silencing [10]. Tobacco plants were developed harboring an antisense construct targeting the AV2 gene of Tomato leaf curl New Delhi Virus (ToLCNDV) [47]. Upon challenging with virus-infectious clones of ToLCNDV, transgenic plants remained asymptomatic, although viral DNA could be detected by PCR. Satyavathi et al. [29] transformed Indian cotton variety F-846 with Agrobacterium transformation by targeting V2 genes of CLCuKoV using a RNAi approach [29]. Transgenic cotton plants exhibited a true Mendelian pattern of inheritance and were tolerant to complex virus including CLCuD. In this study, we are not only reporting the development of cotton transgenic plants, but we have conducted a comprehensive study on the resistance of amplicon V2 RNAi construct. Our results have shown that the amplicon V2 RNAi approach can provide high tolerance in transgenic cotton plants. In order to achieve a resistance to immunity, the amplicon V2 RNAi can be coupled with other types of resistance and tolerance.
Cotton varieties MNH-786 and VH-289 were transformed via Agrobacterium Strain LBA4404. A total of 84 experiments were performed to transform the RNAi gene in cotton. The number of plantlets obtained after 8 weeks for both transgenic varieties was low, i.e., 278 for MNH-786 and 196 for VH-289. Thus, the transformation efficiency of MNH-786 and VH-289 was 3.75 % and 2.88 %, respectively (Table 3). Bakhsh et al. [48] reported higher transformation efficiency (20 %) of Agrobacterium strain (LBA4404) in tobacco plants [48], but cotton is harder to transform, and our results were similar to the work of Majeed et al. [49] in which transformation efficiency in cotton was 5.17 % [49, 50].
Eighty-six putative transgenic plants of MNH-786 and 43 putative transgenic rooted plants of VH-289 were shifted to soil pots. PCR confirmed the successful integration of the RNAi gene into cotton genome. The number of plants was further reduced during acclimatization, and the numbers of T0 generation plants shifted to the field was 24 and 26 for MNH-786 and VH-289, respectively. Once the plants began flowering, their flowers were self-pollinated to avoid gene transfer [51].
The seeds of the T0 generation were sown in the field during August 2013, with ten seeds of a single event sown in a row. The plants were not protected from whitefly attack at this stage.
The whitefly attack on transgenic plants of the T 1 generation was seen at a very early stage of sowing. The plants were continuously observed during this menace. The viral index of all plants in the field was calculated using a formula as described by Akhtar and Khan [34]. Four plants were selected from both varieties (VH-289 and MNH-786) along with one non-transgenic plant to determine the viral titer via RT-PCR. The plants having minimum symptoms showed minimum viral titer and vice versa (Fig. 7). The results of real-time PCR were of utmost importance for the selection of plants for the T 2 generation. The other important factor considered for selection of plants was the PCR result which confirmed RNAi gene integration in T 1 plants. The integration of RNAi gene construct in plants of T 2 generation was confirmed via PCR using gene-specific internal primers. Three out of six MNH-786 and five out of eight VH-289 T3 plants were confirmed after PCR analysis. qPCR assay was performed to quantify virus titer in plants showing minimum and maximum symptoms (from each variety). Interestingly, a positive correlation was observed between the virus titer and disease index.
Both the varieties (MNH-786 and VH-289) showed almost similar results.
To determine the transgene location and copy number in the transgenic plants, the FISH analysis was done. Transformants with a single copy were achieved in both the varieties. In the case of two events from different varieties, the transgene locations were observed on different chromosomes. Transformed plants (vv. MC2-8) showed transgene insertion at chromosome No. 6, whereas in VC2-11 line, it was at chromosome No. 16. This has been observed in other transformation events and the different locations maybe due to various factors which are involved in the transformation of the transgene into the host genome [52, 53].
Conclusion
The molecular analyses performed in this study showed the positive integration of the RNAi gene construct in T 0, T 1, and T 2 generations. Cotton is an important crop not only for Pakistan, but also India where CLCuD is a big threat to cotton as well. Any successful transgenic varieties could have a big impact in countries throughout the globe where CLCuD is a threat. The positive integration of an RNAi-based gene in cotton varieties and absolute quantification determining viral titer of transgenic plants gives hope for a promising future for transgenic crops in Pakistan.
Acknowledgments
This material is based upon work supported by the “Pakistan–U.S. Cotton Productivity Enhancement Program” of ICARDA funded by the United States Department of Agriculture (USDA), Agricultural Research Service (ARS), under Agreement No. 58-6402-0-178F. Any opinions, findings, conclusions, or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the views of the USDA or ICARDA.
Authors’ Contributions
Aneela Yasmeen, Sarfraz Kiani, Afshan Butt, and Aftab Ahmad have done transformation, result analysis, and manuscript preparation. Faheem Akram performed the FISH analysis. Muhammad Zubair, Muhammad Nauman Tahir, and Sohail Akhtar performed the cloning. Imran Amin and Shaheen Aftab developed cloning strategy; Abdul Qayyum Rao and Idrees Ahmad Nasir developed cotton transformation strategy; Tayyab Husnain, Shahid Mansoor, Jodi Scheffler, and Brian Scheffler provided overall supervision for the project and manuscript revision and proofreading.
Funding
The research work was funded by the “Pakistan–U.S. Cotton Productivity Enhancement Program” of ICARDA funded by the United States Department of Agriculture (USDA), Agricultural Research Service (ARS), under Agreement No. 58-6402-0-178F and Grant No. 1198.
Compliance with Ethical Standards
Conflict of Interest
The authors declare no conflict of interests.
Contributor Information
Aneela Yasmeen, Email: anilla_malik5@hotmail.com.
Sarfraz Kiani, Email: frazkiani786@yahoo.com.
Afshan Butt, Email: ashbutt82@gmail.com.
Abdul Qayyum Rao, Email: qayyumabdul77@yahoo.com.
Faheem Akram, Email: faheem_bot@yahoo.com.
Aftab Ahmad, Email: warraich6229@cemb.edu.pk.
Idrees Ahmad Nasir, Email: dr.idrees@gmail.com.
Tayyab Husnain, Email: tayyabhusnain@yahoo.com.
Shahid Mansoor, Email: shahidmansoor7@gmail.com.
Imran Amin, Email: imranamin1@yahoo.com.
Shaheen Aftab, Email: sasad@nibge.org.
Muhammad Zubair, Email: zubair.nibge@gmail.com.
Muhammad Nouman Tahir, Email: nouman.nibge@yahoo.com.
Sohail Akhtar, Email: m.sohail.awan@hotmail.com.
Jodi Scheffler, Email: jodi.scheffler@ars.usda.gov.
Brian Scheffler, Email: Brian.Scheffler@ars.usda.gov.
References
- 1.Azam S, Samiullah TR, Yasmeen A, Din S, Iqbal A, Rao AQ, et al. Dissemination of Bt cotton in cotton growing belt of Pakistan. Advancements in Life Sciences. 2013;1:18–26. [Google Scholar]
- 2.Mansoor S, Amrao L, Amin I, Briddon RW, Malik K, et al. First report of cotton leaf curl disease in central and southern Sindh province in Pakistan. Plant Disease. 2006;90:826. doi: 10.1094/PD-90-0826A. [DOI] [PubMed] [Google Scholar]
- 3.Shahid AA, Bano S, Khalid S, Samiullah TR, Bajwa KS, Ali MA. Biosafety assessment of transgenic Bt cotton on model animals. Advancements in Life Sciences. 2016;3:97–108. [Google Scholar]
- 4.Rahman M, Hussain D, Malik TA, Zafar Y. Genetics of resistance against cotton leaf curl disease in Gossypium hirsutum. Plant Pathology. 2005;54:764–772. doi: 10.1111/j.1365-3059.2005.01280.x. [DOI] [Google Scholar]
- 5.Mansoor S, Amin I, Iram S, Hussain M, Zafar Y, et al. The breakdown of resistance in cotton to cotton leaf curl disease in Pakistan. Plant Pathology. 2003;52:784. doi: 10.1111/j.1365-3059.2003.00893.x. [DOI] [Google Scholar]
- 6.Amrao L, Amin I, Shahid MS, Briddon RW, Mansoor S. Cotton leaf curl disease in resistant cotton is associated with a single begomovirus that lacks an intact transcriptional activator protein. Virus Research. 2010;152:153–163. doi: 10.1016/j.virusres.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 7.Akhtar KP, Haq MA, Hussain M, Khan AI. Whitefly transmitted gemini virus and associated disorder in cotton: A review. Pakistan Journal of Phytopathology. 2002;14:140–150. [Google Scholar]
- 8.Briddon RW, Markham PG. Cotton leaf curl virus disease. Virus Research. 2001;71:151–159. doi: 10.1016/S0168-1702(00)00195-7. [DOI] [PubMed] [Google Scholar]
- 9.Beck EH, Fettig S, Knake C, Hartig K, Bhattarai T. Specific and unspecific responses of plants to cold and drought stress. Journal of Biosciences. 2007;32:501–510. doi: 10.1007/s12038-007-0049-5. [DOI] [PubMed] [Google Scholar]
- 10.Amin I, Hussain K, Akbergenov R, Yadav JS, Qazi J, et al. Suppressors of RNA silencing encoded by the components of the cotton leaf curl begomovirus-betasatellite complex. Molecular Plant Microbe Interactions. 2011;24:973–983. doi: 10.1094/MPMI-01-11-0001. [DOI] [PubMed] [Google Scholar]
- 11.Amin I, Mansoor S, Amrao L, Hussain M, Irum S, et al. Mobilization into cotton and spread of a recombinant cotton leaf curl disease satellite. Archives of Virology. 2006;151:2055–2065. doi: 10.1007/s00705-006-0773-4. [DOI] [PubMed] [Google Scholar]
- 12.Kauter G. Cotton leaf curl virus study tour. Narrabri, NSW: Australian Cotton Growers Research Association Inc; 2007. [Google Scholar]
- 13.Khan MAU, Shahid AA, Rao AQ, Shahid N, Latif A, ud Din S, et al. Defense strategies of cotton against whitefly transmitted CLCuV and Begomoviruses. Advancements in Life Science. 2015;2:58–66. [Google Scholar]
- 14.Aftab B, Shahid MN, Riaz S, Jamal A, Mohamed BB, et al. Identification and expression profiling of CLCuV-responsive transcripts in upland cotton (Gossypium hirsutum L.) Turkish Journal of Biology. 2014;38:226–237. doi: 10.3906/biy-1307-55. [DOI] [Google Scholar]
- 15.Khatoon S, Kumar A, Sarin NB, Khan JA. RNAi-mediated resistance against cotton leaf curl disease in elite Indian cotton (Gossypium hirsutum) cultivar Narasimha. Virus Genes. 2016;52(4):530–537. doi: 10.1007/s11262-016-1328-8. [DOI] [PubMed] [Google Scholar]
- 16.Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruiz-Ferrer V, Voinnet O. Roles of plant small RNAs in biotic stress responses. Annual Review of Plant Biology. 2009;60:485–510. doi: 10.1146/annurev.arplant.043008.092111. [DOI] [PubMed] [Google Scholar]
- 18.Hannon GJ. RNA Interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 19.Baulcombe D. RNA silencing. Trends in Biochemistry Sciences. 2005;30:290. doi: 10.1016/j.tibs.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 20.Helliwell CA, Waterhouse PM, Engelke DR, Rossi JJ. Constructs and methods for Hairpin RNA-mediated gene silencing in plants. Methods in Enzymology. 2005;392:24. doi: 10.1016/S0076-6879(04)92002-2. [DOI] [PubMed] [Google Scholar]
- 21.Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- 22.Voinnet O. RNA silencing as a plant immune system against viruses. Trends in Genetics. 2001;17:449–459. doi: 10.1016/S0168-9525(01)02367-8. [DOI] [PubMed] [Google Scholar]
- 23.Zha W, Peng X, Chen R, Du B, Zhu L, et al. Knockdown of midgut genes by dsRNA-transgenic plant-mediated RNA interference in the hemipteran insect Nilaparvatalugens. PLOS One. 2011;6(5):e20504. doi: 10.1371/journal.pone.0020504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mao YB, Tao XY, Xue XY, Wang LJ, Chen XY. Cotton plants expressing CYP6AE14 double-stranded RNA show enhanced resistance to bollworms. Transgenic Research. 2011;20:665–673. doi: 10.1007/s11248-010-9450-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vanderschuren H, Stupak M, Fütterer J, Gruissem W, Zhang P. Engineering resistance to geminiviruses–review and perspectives. Plant Biotechnology Journal. 2007;5:207–220. doi: 10.1111/j.1467-7652.2006.00217.x. [DOI] [PubMed] [Google Scholar]
- 26.Shepherd DN, Martin DP, Thomsonm JA. Transgenic strategies for developing crops resistant to geminiviruses. Plant Science. 2009;176:1–11. doi: 10.1016/j.plantsci.2008.08.011. [DOI] [Google Scholar]
- 27.Aragao FJL, Faria JC. First transgenic geminivirus-resistant plant in the field. Nature Biotechnology. 2009;227:1086–1088. doi: 10.1038/nbt1209-1086. [DOI] [PubMed] [Google Scholar]
- 28.Aragao FJ, Nogueira EO, Tinoco ML, Faria JC. Molecular characterization of the first commercial transgenic common bean immune to the Bean golden mosaic virus. Journal of Biotechnology. 2013;166:42–50. doi: 10.1016/j.jbiotec.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 29.Satyavathi VV, Prasad V, Kirthi N, Maiya SP, Savithri HS, et al. Development of cotton transgenics with antisense AV2 gene for resistance against cotton leaf curl virus (CLCuD) via Agrobacterium tumefaciens. Plant Cell Tissue Organ Culture. 2005;81:55–63. doi: 10.1007/s11240-004-2777-7. [DOI] [Google Scholar]
- 30.Xie Y, Liu Y, Meng M, Chen L, Zhu Z. Isolation and identification of a super strong plant promoter from cotton leaf curl Multan virus. Plant Molecular Biology. 2003;53:1–14. doi: 10.1023/B:PLAN.0000009257.37471.02. [DOI] [PubMed] [Google Scholar]
- 31.Rao AQ, Bakhsh A, Kiani S, Shahzad K, Shahid AA, Husnain T, Riazuddin S. The myth of plant transformation. Biotechnology Advances. 2009;27:753–763. doi: 10.1016/j.biotechadv.2009.04.028. [DOI] [PubMed] [Google Scholar]
- 32.Rao AQ, Irfan M, Saleem Z, Husnain T, Riazuddin S. Phytochrome B gene of Arabidopsis thaliana increases the yield of cotton (Gossypium hirsutum) Journal of Zhejiang University Science B. 2011;12(4):326–334. doi: 10.1631/jzus.B1000168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bullentin. 1987;19:11–15. [Google Scholar]
- 34.Akhtar KP, Khan MSI. Modified scale for the assessment of cotton leaf curl virus (CLCuV) Pakistan Journal of Phytopathology. 2002;14:88–90. [Google Scholar]
- 35.Farooq, et al. An overview of cotton leaf curl virus disease (CLCuD) a serious threat to cotton productivity. Australian Journal of Crop Science. 2011;5(13):1823–1831. [Google Scholar]
- 36.Akhtar KP, Khan AI, Hussain M, Haq MA, Khan MSI. Upland cotton varietal response to cotton leaf curl virus (CLCuV) Tropical Agricultural Research and Extension. 2003;5:29–34. [Google Scholar]
- 37.Southern EM. Detection of specific sequence among DNA fragments separated by gel electrophoresis. Journal of Molecular Biology. 1975;98:503–517. doi: 10.1016/S0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 38.Rahman M, Noreen S, Husnain T, Riazuddin S. Fast and efficient method to determine the position of alien genes in transgenic plants. Emirates Journal of Food and Agriculture. 2010;22(3):223–231. [Google Scholar]
- 39.Seal SE, Jeger MJ, Van den Bosch F. Begomovirus evolution and disease management. Advances in Virus Research. 2006;67:297–316. doi: 10.1016/S0065-3527(06)67008-5. [DOI] [PubMed] [Google Scholar]
- 40.Mansoor S, Briddon RW, Zafar Y, Stanley J. Geminivirus disease complexes an emerging threat. Trends in Plant Science. 2003;8:128–134. doi: 10.1016/S1360-1385(03)00007-4. [DOI] [PubMed] [Google Scholar]
- 41.Aragão FJL, Ribeiro SG, Barros LMG, Brasileiro AM, Maxwell DP, et al. Transgenic beans (Phaseolus vulgaris L.) engineered to express viral antisense RNAs show delayed and attenuated symptoms to bean golden mosaic geminivirus. Molecular Breeding. 1998;4:491–499. doi: 10.1023/A:1009613607559. [DOI] [Google Scholar]
- 42.Ali I, Amin I, Briddon RW, Mansoor S. Artificial microRNA-mediated resistance against the monopartite begomovirus cotton leaf curl Burewala virus. Virology Jounral. 2013;10:231. doi: 10.1186/1743-422X-10-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ammara UE, Mansoor S, Saeed M, Amin I, Briddon RW, et al. RNA interference-based resistance in transgenic tomato plants against tomato yellow leaf curl virus-Oman (TYLCV-OM) and its associated betasatellite. Virology Journal. 2015;12:38. doi: 10.1186/s12985-015-0263-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nahid N, Amin I, Briddon RW, Mansoor S. RNA interference based resistance against a legume Mastrevirus. Virology Journal. 2011;8:499. doi: 10.1186/1743-422X-8-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mubin M, Amin I, Amrao L, Briddon RW, Mansoor S. The hypersensitive response induced by the V2 protein of a monopartite begomovirus is countered by the C2 protein. Molecular Plant Pathology. 2010;1:245–254. doi: 10.1111/j.1364-3703.2009.00601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amin I, Patil BL, Briddon RW, Mansoor S, Fauquet CM. Comparison of phenotypes produced in response to transient expression of genes encoded by four distinct begomoviruses in Nicotianabenthamiana and their correlation with the levels of developmental miRNAs. Virology Journal. 2011;8:238. doi: 10.1186/1743-422X-8-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mubin M, Mansoor M, Hussain M, Zafar Y. Silencing of the AV2 gene by antisense RNA protects transgenic plants against a bipartite begomovirus. Virology Journal. 2007;4:10. doi: 10.1186/1743-422X-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bakhsh A, Anayol E, Ozcan SF. Comparison of transformation efficiency of five Agrobacterium tumefaciens strains in Nicotianatabacum L. Emirates Journal of Food and Agriculture. 2014;26(3):259–264. [Google Scholar]
- 49.Majeed A, Husnain T, Riazuddin S. Transformation of virus- resistant genotype of Gossypium hirsutum L. with pesticidal gene. Plant Biotechnology. 2000;17(2):105–110. doi: 10.5511/plantbiotechnology.17.105. [DOI] [Google Scholar]
- 50.McCabe DE, Martinell BJ. Transformation of elite cotton cultivars via particle bombardment of meristems. Nature Biotechnology . 1993;11:596–598. doi: 10.1038/nbt0593-596. [DOI] [Google Scholar]
- 51.Umbeck P, Swain W, Yang N. Inheritance and expression of genes for kanamycin and chloramphenicol resistance in transgenic cotton plants. Crop Science. 1989;29(1):196–201. doi: 10.2135/cropsci1989.0011183X002900010042x. [DOI] [Google Scholar]
- 52.Kohli A, Gahakwa D, Vain P, Laurie DA, Christou P. Transgene expression in rice engineered through particle bombardment: Molecular factors controlling stable expression and transgene silencing. Planta. 1999;208:88–97. doi: 10.1007/s004250050538. [DOI] [Google Scholar]
- 53.Yin Z, Malepszy S. The transgenes are expressed with different level in plants. Biotechnologia. 2003;2(61):236–260. [Google Scholar]