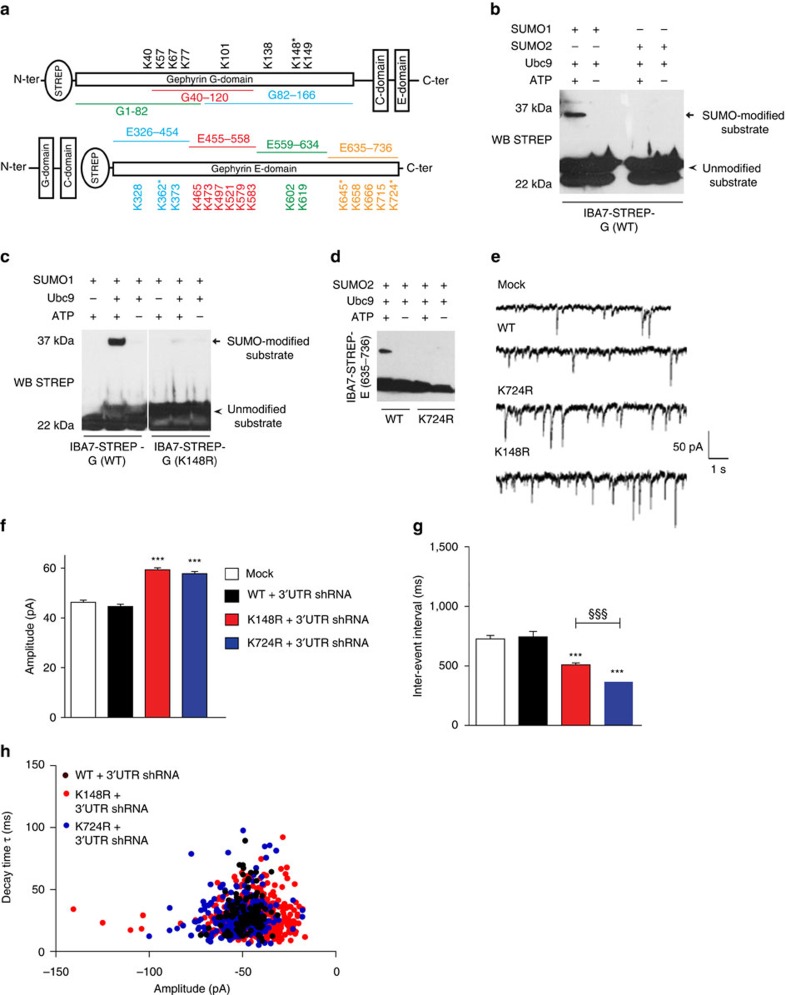
Figure 2. Identification and characterization of gephyrin SUMO sites.
(a) Cartoon of gephyrin G-domain and E-domain peptide sequences with identified surface-exposed lysine residues. The identified SUMO1 and SUMO2 conjugation sites are marked *. (b) In vitro SUMOylation using bacterial expressed and purified STREP-G domain shows SUMO-1 conjugation but not SUMO2 conjugation. (c) In vitro SUMOylation using STREP-G (K148R) mutant abolishes SUMO-1 conjugation. (d) In vitro SUMO2 reactions using STREP-E (635–736) and STREP-E (635–736) K724R mutant shows SUMO2 conjugation in the WT peptide but SUMO2 conjugation is completely abolished in the K724R mutant peptide. (e) Sample mIPSC trace measurements in neurons transfected with eGFP–K148R, eGFP–K724R along with gephyrin 3′-UTR shRNA compared with WT or mock transfected neurons. (f) Average mIPSC amplitude in neurons transfected with eGFP–K148R, eGFP–K724R along with gephyrin 3′-UTR shRNA compared with WT or mock-transfected shows significantly increased amplitude. (g) Average mIPSC inter-event interval in neurons transfected with eGFP–K148R and eGFP–K724R along with gephyrin 3′-UTR shRNA compared with WT or mock-transfected shows significantly reduced inter-event intervals. (h) Decay kinetics of mIPSC currents in WT, K148R- and K724R-transfected neurons. Images from four independent experimental replicates were analysed; error bars are s.e.m.