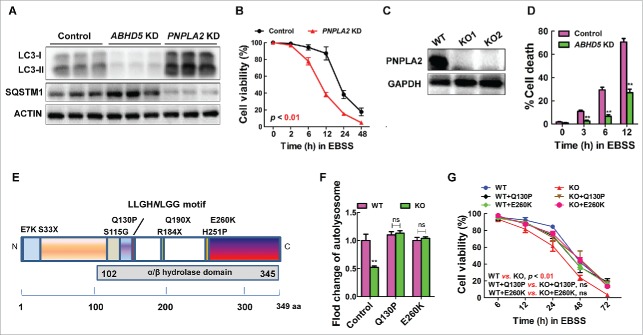
Figure 4.
ABHD5 regulates autophagic flux independent of its canonical metabolic activity. (A) Western blots of autophagy-related proteins (LC3-I, LC3-II, SQSTM1) in ABHD5-silenced, PNPLA2-silenced and control CCD841CON cells 24 h following exposure to EBSS. (B) PNPLA2-silenced and control CCD841CON cells were treated with EBSS for 24 h, and the cell viability was determined by MTT assay. (C) Western blots of PNPLA2 in control and CRISPR/Cas9-mediated PNPLA2 knockout (KO) CCD841CON cells. (D) ABHD5 expression was silenced in PNPLA2 knockout CCD841CON cells, and the percentage of dead cells was determined at the indicated time points by trypan blue exclusion assay. (E) Predicted domain structure of ABHD5, including the α/β hydrolase domain. The sites of mutations are indicated. (F) ABHD5+/+ (WT) and ABHD5−/− (KO) CCD841CON cells were transfected with mutated ABHD5 (Q130P and E260K) or control vector plasmids. Statistical analysis of the number of autophagosomes/cell in different groups (a random number of 30 cells were selected for each group). (G) ABHD5+/+ (WT) and ABHD5−/− (KO) CCD841CON cells were transfected with mutated ABHD5 (Q130P and E260K) or control vector plasmids. Cell viability of cells at different time points (6, 12, 24, 48, 72 h) after the exposure to EBSS was measured by MTT assay. (**, p < 0.001; ns, no significance).