Fig. 3.
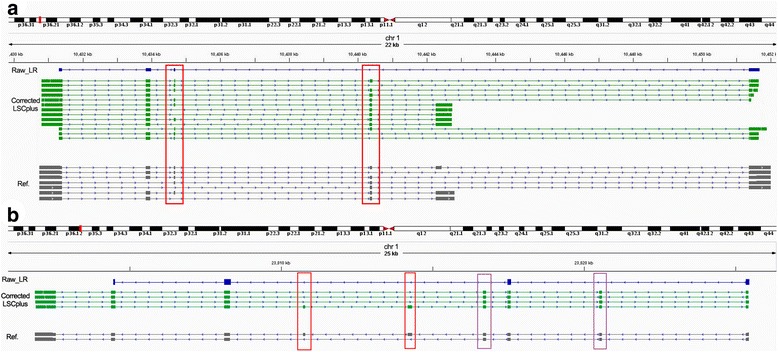
Two local details of the alignments of raw LRs and LSCplus-corrected LRs. (a) Two positions of isoform identification. (b) Two positions of isoform identification and Two positions of exons recovery. Error correction of RNA-seq data provides more accurate mapping of transcripts. A genome browser view of transcriptome alignments using uncorrected (blue) and corrected (green) PacBio reads. Color blocks represent the exons. Before correction, only one potential transcript isoform was detected with any exons missing (indicated with purple rectangles), and after correction, the corrected sequences matched the reference annotations end to end with no exons missing. As a result, the isoforms (indicated with red rectangles) at the displayed reference locus in the reference annotation were confirmed by corrected PacBio RNA-seq reads
