Fig. 1.
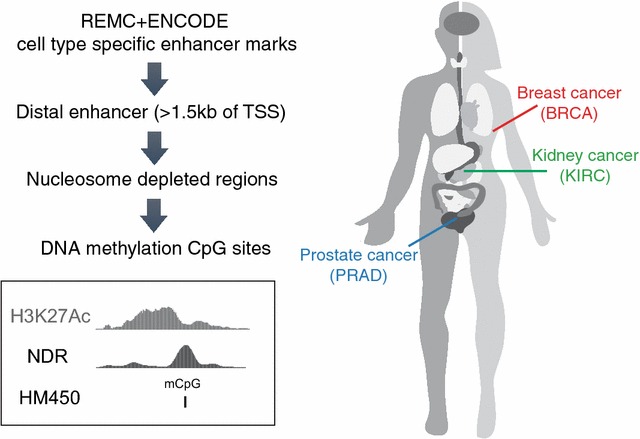
Study design. To define genomic regions for analysis of enhancer activity in tumor samples, we used the genomic coordinates of enhancers identified by REMC and ENCODE for 98 tissues or cell lines, plus genomic coordinates of additional H3K27Ac ChIP-seq peaks from several cancer cell lines and normal cells for breast, prostate, and kidney. We then selected the subset of these regulatory elements that are located >1.5 kb from a known transcription state site (TSS), as defined using GENCODE v19. We further narrowed the regions by intersecting with the set of ENCODE Master DNaseI-seq peaks from 125 tissues or cell lines or DNaseI-seq, FAIRE-seq, or NOMe-seq peaks of corresponding cell types (Additional file 2: Table S1). The HM450 array probes that overlapped the narrowed enhancer regions were then used to study enhancer activity in normal and tumor tissues
