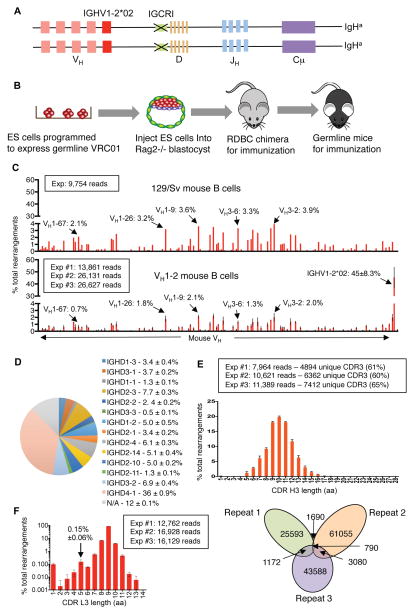
Figure 1. Generation and characterization of VH1-2 mouse model.
(A) Illustration of genetic modifications in the VH1-2 mouse mode (see text, Figure S1A and S1B for details). (B) Illustration of Rag2 deficient blastocyst complementation (See text for details). (C) HTGTS-rep-seq analysis of VH usage in VH1-2 mouse model and in control 129/Sv mouse. The X-axis represents VH locus from the distal to the D-proximal ends; a subset of mouse VHs are labeled for comparison between 129/Sv control and VH1-2 mice. The histogram displays the percent usage of each VH of all productive VH(D)JH rearrangements. Data for the VH1-2 mouse model were average of three experiments with error bars representing standard deviations; data for control 129/Sv mouse is consistent with prior studies (Lin et al. 2016). (D) Pie chart Illustration of D segment usage in productive IGHV1-2*02 rearrangements in VH1-2 mouse model, average D usage frequency with standard deviation for three biological repeats is shown in adjacent panel. (E) Length distribution of IGHV1-2*02 CDR H3 in VH1-2 mouse model derived from data in (C). The number of total reads and unique reads for IGHV1-2*02 associated CDR H3’s are shown and Venn diagram, which reveals tremendous CDR3 complexity since there is little overlap in CDR H3 sequences in three technical repeats of a single B cell sample (see Lin et al. 2016). (F) Length distribution of CDR L3 of mouse IgL chains in VH1-2 mice based on three biological replicates with error bars displayed and frequency of 5-amino acid CDR L3 indicated. Other details are in Methods.