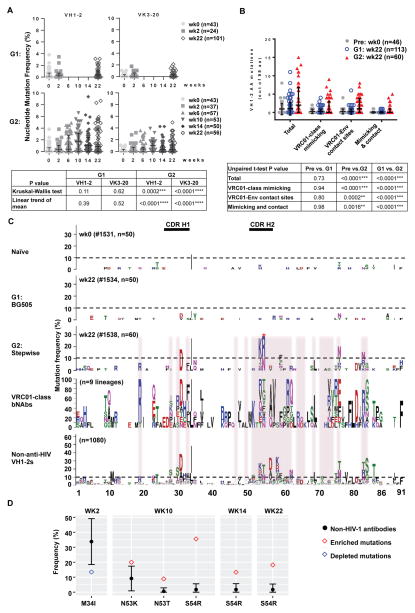
Figure 5. Stepwise immunization of VH1-2/LC mice elicited VRC01-class CD4 binding site-specific antibodies with increased SHM.
(A) Nucleotide mutation frequency in paired VH1-2 and VK3-20 genes, derived from CD4bs-specific IgG+ B cells of the BG505-immunized mice (G1) and stepwise-immunized mice (G2). Each dot represents one VH1-2 or VK3-20 chain in a VH1-2/VK3-20 paired antibody (n, number of VH1-2/VK3-20 pairs in each animal at indicated time points). The median with interquartile range is plotted. (B) Numbers of total, VRC01-class mimicking, Env-contacting, and both mimicking and contacting amino acid mutations in each VH1-2 HC amplified from specified mice plotted with median and interquartile range and statistically assessed using an unpaired t-tests. VRC01-class mimicking mutations are present in at least two of 9 published VRC01-class bnAb lineages. Env-contacting sites are based on VRC01-gp120 or gp160 crystal structures. (C) Amino acid mutations in all VH1-2 HCs from each specified mouse shown in sequence logo profiles. “n” is the number of all VH1-2 chains amplified from each animal. For reference, 9 published VRC01-class HC lineages are represented below panel G2. VRC01-envelope contact sites are highlighted in pink. The mutation profile of IGHV1-2 constructed from 1080 non-HIV-1 neutralizing antibody lineages (from three healthy donors) is also shown. The red asterisk marks significantly enriched S54R mutations in the G2:wk22 sample. (D) Enriched and depleted amino acid substitutions in VH1-2 cloned from the sequential immunization group G2 compared to non-HIV-1 neutralizing antibodies. The calculated mean occurrence and standard deviations of each mutation and 95% CI are depicted in black. The frequency of enriched (red diamond) or depleted (blue diamond) mutations locates outside of the calculated 95% CI.