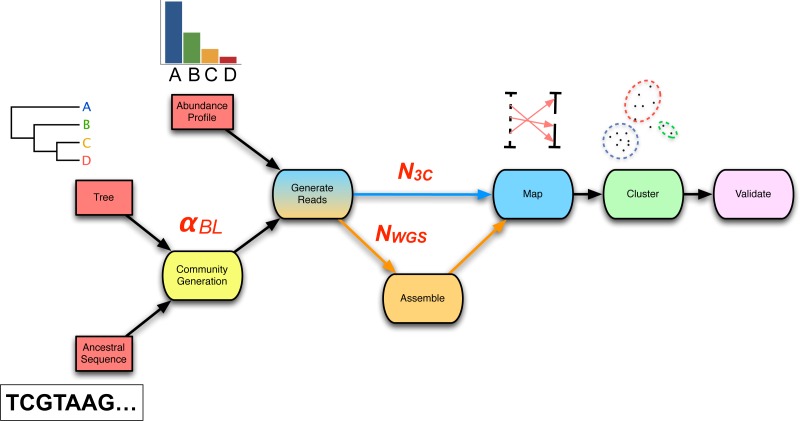
Figure 2. The 3C sequencing simulation pipeline used within the parameter sweep.
An ancestral sequence and phylogenetic tree are used in simulating a process of genome evolution with varying divergence (αBL). The resulting evolved genomes are subsequently subjected to in silico high-throughput sequencing, producing both WGS and 3C read-sets of chosen depth (NWGS, N3C). WGS reads are assembled and 3C read-pairs are mapped to the resulting contigs to generate a 3C-contig graph. Finally, the graph is supplied to a clustering algorithm and the result validated against the relevant gold-standard.