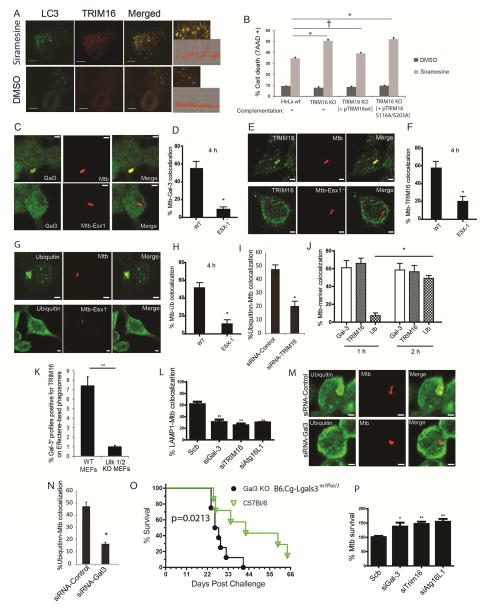
Figure 7. TRIM16-Galectin-3 system protects against endomembrane damage associated with lysosomal dysfunction and pathogen-mediated phagosomal perforation.
(A) Confocal images of HeLa cells treated with DMSO or siramesine; immunofluorescence, endogenous LC3B and TRIM16 revealed with corresponding antibodies. Graphs on the right, line tracing colocalization analyses. (B) HeLa, CRISPR TRIM16KO HeLa cells A9, or TRIM16KO HeLa cells A9 transfected with expression plasmids for wt TRIM16 or TRIM16S116A/S203A mutant were incubated with siramesine and cell death was measured using 7AAD nuclear staining. Data, means; n>3; †, p ≥ 0.05 *, p < 0.05 (ANOVA). (C,D) RAW264.7 macrophages were infected with Alexa-568-labeled wild-type M. tuberculosis Erdman or its ESX-1 mutant at MOI=10 for 4 h and then processed for confocal microscopy analysis of M. tuberculosis (Mtb) colocalization with Galectin-3. Data, means ± SEM (n>3; at least 100 Mtb phagosomes per condition were quantified); *, p<0.05 (t-test). Bar 2 μm. (E,F) RAW264.7 macrophages were infected with Alexa-568-labeled wild-type Mtb Erdman (WT) or its ESX-1 mutant at MOI=10 for 4 h and then processed for confocal microscopy analysis for the colocalization of Mtb with TRIM16. Data and statistics as in E. (G,H) RAW264.7 cells were infected with Alexa-568-labeled Mtb wild-type Erdman or its ESX-1 mutant for 4 h and then processed for immunofluorescence staining with anti-ubiquitin. Data and statistics as in D. (I) RAW264.7 macrophages were transfected with siRNAs against TRIM16 or control siRNAs for 48 h. Cells were then infected with Alexa-568-labeled wild-type Mtb Erdman for 4 h and processed for confocal microscopy analysis for the colocalization of Mtb with ubiquitin. Data and statistics as in D. (J) Time course of marker appearance on Mtb phagosomes; RAW264.7 macrophages were infected with Alexa-568-labeled wild-type M. tuberculosis Erdman at MOI=10 for 1 and 2 h and processed and data analyzed as in panels C–H. (K) Effectene-coated beads were phagocytosed by MEFs in 96 well plates, incubated for up to 24 h (> 3h), stained with antibodies, and processed for HC microscopy (see images in Figure S7D). Data: means (n>3); t-test *, p < 0.05. Ninety six-well plates, with >12 wells/condition with >500 valid objects/well. (L) RAW264.7 cells were transfected with siRNAs against TRIM16, Galectin-3, or ATG16L1 or control siRNAs for 48 h. Cells were then infected with Alexa-568-labeled wild-type Mtb Erdman for 4 h followed by immunofluorescence staining for LAMP1 and % colocalization of Mtb phagosomes with LAMP1 determined (for representative images see Figure S7G). Data, means ± SEM (n=3; at least 100 Mtb phagosomes per condition were quantified). (M,N) RAW264.7 cells were transfected with siRNAs against Galectin-3 or control siRNAs for 48 h. Cells were then infected with Alexa-568-labeled wild-type Mtb Erdman for 4 h followed by immunofluorescence staining for ubiquitin. Data and statistics as in L. (O) Survival of wt C57BL and Galectin-3 C57BL knockout mice in a short-term acute infection model (high dose; 1-3xe3 CFU) with M. tuberculosis Erdman aerosols. For data from a chronic model of infection with lower doses of M. tuberculosis, see Figure S7J,K). (P) RAW264.7 cells were transfected with siRNAs against TRIM16, Galectin-3, or ATG16L1 or control siRNAs for 48 h. Cells were then infected with wild-type Mtb Erdman for 1 h (t=0) at MOI=10. Cells were then washed three times with complete medium to remove uninternalized mycobacteria and then continued to grow in complete medium for another 24 h (t=24). Cells were then harvested for CFU analysis of Mtb intracellular survival. Data, means ± SEM of CFU at t=24 normalized to CFU at t=0 (n>3 independent experiments). *p < 0.05 and **p < 0.01, t-test relative to the scrambled (control) siRNA set at 100%.