Abstract
Background
Cognitive dysfunction has been recognized as a cardinal feature of schizophrenia. Elucidating the neurobiological substrates of cognitive dysfunction in schizophrenia would help identify the underlying mechanism of this disorder. The rs1064395 single nucleotide polymorphism, within the gene encoding neurocan (NCAN), is reported to be associated with schizophrenia in European populations and may influence brain structure in patients with schizophrenia.
Methods
In this study, we aimed to explore whether NCAN rs1064395 confers some risk for schizophrenia and cognitive dysfunction in Han Chinese. We recruited 681 patients with schizophrenia and 699 healthy subjects. Two hundred and fifty-four patients were evaluated according to Repeatable Battery for the Assessment of Neuropsychological Status (RBANS).
Results
There were no significant differences in genotype or allele distributions of the rs1064395 polymorphism between the schizophrenia and control groups. Patients showed significantly poorer performance than controls on immediate memory, visuospatial skill, language, attention, delayed memory, and total RBANS score. Patients with the A/A or A/G genotype of rs1064395 had lower scores of immediate memory, visuospatial skill, attention, and total RBANS score than those with the G/G genotype. We performed an expression quantitative trait loci analysis and observed a significant association between rs1064395 and NCAN expression in the frontal (P=0.0022, P=0.022 after Bonferroni correction) and cerebellar cortex (P=0.0032, P=0.032 after Bonferroni correction).
Conclusion
Our findings indicate that this single nucleotide polymorphism may be a risk factor for cognitive dysfunction in patients with schizophrenia. Further investigations are warranted for validation purposes and to identify the precise mechanism by which rs1064395 influences cognitive performance in patients with schizophrenia.
Keywords: NCAN, schizophrenia, cognitive function, polymorphism, eQTL
Introduction
Schizophrenia is a severe and chronic psychiatric disorder that affects ~1% of the population worldwide. Although its pathophysiological mechanism remains unclear, there is compelling evidence from family, twin and adoption studies supporting the involvement of a genetic predisposition to schizophrenia, with an estimated heritability up to 80%. While multiple susceptibility loci for schizophrenia diagnosis have been identified, only few have been widely replicated. One suggestion for such a dilemma is that novel phenotypes be defined to reflect neurobiological processes in schizophrenia.1
Cognitive dysfunction is recognized as a cardinal feature of schizophrenia that implicates functional outcome.2,3 There is a realm of evidence showing that patients with schizophrenia exhibit impairment across a range of cognitive domains, such as working memory, language function, executive function, episodic memory, processing speed, attention, inhibition, and sensory processing.4 The neurodevelopmental hypothesis of schizophrenia is therefore based, at least in part, on the presence of cognitive dysfunction in schizophrenic patients.5 Therefore, elucidating the neurobiological substrates of cognitive dysfunction in schizophrenia would help to identify the underlying mechanism of this disorder.
Neurocan is a member of the lectican/chondroitin sulfate proteoglycan protein family and consists of a neurocan core protein and chondroitin sulfate. Neurocan is an important constituent of the brain extracellular matrix that is involved in neural crest cell migration, neurite outgrowth, neuronal cell adhesion, and synaptic plasticity.6 The gene encoding neurocan (NCAN) is located on chromosome 19p13 and has been reported to be a genetic susceptibility region for schizophrenia.7 A recent meta-analysis employed three independent studies and showed a single nucleotide polymorphism (SNP), rs1064395, in the 3′-untranslated region (3′-UTR) of the NCAN gene that is significantly associated with schizophrenia in European populations.8 To describe the effect of rs1064395 on brain structure, Schultz et al9 performed an imaging genetics study and found an implication of this SNP with increased cortical folding (a putative marker of increased neuronal efficiency) in the occipital and prefrontal cortex, but not in healthy subjects, which may establish schizophrenia susceptibility during neurodevelopment. Taken together, the NCAN rs1064395 may be a functional polymorphism that influences the brain structure that results in neurodevelopmental risk for schizophrenia.
To the best of our knowledge, no genetic study to date has addressed the association between NCAN and schizophrenia in Han Chinese. Building on this premise, we aimed to explore whether NCAN rs1064395 confers some risk of susceptibility to schizophrenia among a Chinese Han population. Given the important neurobiological function of NCAN, we subsequently aimed to determine whether the rs1064395 polymorphism influences cognitive function in patients with schizophrenia.
Methods
Subjects
We recruited 681 patients with schizophrenia from four mental hospitals in Eastern China, including Shanghai Mental Health Center, Tongde Hospital of Zhejiang Province, Jinhua Second Hospital, and Wenzhou Kangning Hospital. All patients met the diagnoses of schizophrenia according to the Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition (DSM-IV) and had no other observable physical disease or other psychiatric disorder aside from schizophrenia. Among them, 254 schizophrenic patients were used to test cognitive function, whose inclusion criteria were based on our previous publications:10,11 1) duration of illness <5 years; 2) has a minimum education of primary middle school; 3) receives atypical antipsychotic monotherapy; 4) has maintained a stable condition for >6 months before entry into the study; and 5) has a positive and negative syndrome scale total score <60.
Six hundred and ninety-nine healthy controls were recruited from hospital staff and students of School of Medicine in Shanghai, and then interviewed by a specialized psychiatrist using the Structured Clinical Interview for DSM-IV-TR Axis I Disorders – Patient Edition. Aside from the controls enrolled for the genetic analysis, we recruited 72 healthy subjects undergoing cognitive evaluation from the Shanghai region.
All procedures for this study were reviewed and approved by the Institutional Review Boards of Shanghai Mental Health Center and other participating institutions. This study was performed in strict accordance with the Declaration of Helsinki and other relevant national and international regulations. Written informed consent was obtained from each participant prior to any procedures related to this study being performed.
Evaluation
The Repeatable Battery for the Assessment of Neuropsychological Status (RBANS) was the primary outcome instrument used in this study.12 The 12-item RBANS consists of five subsets, corresponding to five domains of the neuropsychological process: 1) immediate memory (list learning and story memory); 2) visuospatial/constructional (figure copy and line orientation); 3) language (picture naming and semantic fluency); 4) attention (digit span and coding); and 5) delayed memory (list learning free recall, list learning recognition, story memory free recall, and figure free recall). RBANS has good validity and reliability in Chinese people13 and works well in genetic studies on schizophrenia.14–16
Genotyping
Genomic DNA from all participants was extracted from peripheral blood using a Tiangen DNA Isolation Kit (Tiangen Biotech, Beijing, People’s Republic of China). Genotyping of the rs1064395 polymorphism was carried out according to the methods described in our previous studies.17,18 Table S1 details the primer information. Briefly, polymerase chain reaction (PCR) amplification was performed in a volume of 25 μL containing one primer pair for each SNP. PCR primers were also used for sequencing. Sequencing results were analyzed using DNAStar package (DNAStar Inc., Madison, WI, USA), and the original sequencing chromatograms of each sample were checked manually.
Psychiatric genomics consortium data analysis
To validate the association between the rs1064395 polymorphism and schizophrenia, we extracted schizophrenia genetic association data from the Psychiatric Genomics Consortium (PGC, http://www.broadinstitute.org/mpg/ricopili/) database19 and reanalyzed the data set as an independent sample.
Brain expression quantitative trait loci analysis
Schizophrenia originates from structural and functional brain abnormalities,20 and brain samples are presumably appropriate for expression quantitative trait loci (eQTL) analysis of risk SNP(s).21,22 Here, we performed an eQTL analysis to determine whether the rs1064395 polymorphism is associated with NCAN expression in the brain using the brain eQTL database (http://caprica.genetics.kcl.ac.uk/BRAINEAC/), a large exon-specific eQTL data set covering ten human brain regions. More detailed information can be found in the original study.23
Statistical analysis
Demographic variables between case and control groups were compared by use of independent t-tests for quantitative variables and Fisher’s exact test for qualitative variables. Analysis of covariance (ANCOVA) was used to compare RBANS scores between case and control groups, controlling for demographic characteristics. Hardy–Weinberg equilibrium testing, and allele and genotype frequency analyses were conducted using SHEsis (http://analysis.bio-x.cn).24 SNPStats (http://bioinfo.iconcologia.net/snpstats/start.htm) was used to examine the association between rs1064395 and schizophrenia under four inheritance models, including codominant, dominant, recessive, and log-additive models.25 The possible effects of the rs1064395 genotypes on cognitive function were examined using ANCOVA by comparing the mean RBANS scores of each genotype. Variables that affect cognitive parameters (ie, age, gender, education, and duration of illness) were included as covariates. Statistical analyses were performed using SPSS 17.0 (SPSS Inc., Chicago, IL, USA). All P-values were two-tailed, and those <0.05 were considered statistically significant after Bonferroni correction.
Results
The genotypic distribution of the rs1064395 polymorphism in the control group was in accordance with Hardy–Weinberg equilibrium. The results from single marker analysis are shown in Table 1. There were no significant differences in genotype or allele distributions of the rs1064395 polymorphism between the schizophrenia and control groups. Unconditional logistic regression was performed to further detect an association between rs1064395 and schizophrenia after adjusting for age and gender. We did not observe any significant association under four inheritance models (Table S2). Next, we extracted the schizophrenia genetic association data from the PGC database and observed a significant association between rs1064395 and schizophrenia (P=0.000256) (Figure S1).
Table 1.
Distribution of rs1064395 genotype and allele in schizophrenia patients and controls
| SNP rs1064395 | N | Genotype, n (%)
|
P-value | Allele, n (%)
|
P-value | OR (95% CI) | P-valuea | |||
|---|---|---|---|---|---|---|---|---|---|---|
| A/A | G/A | G/G | A | G | ||||||
| Case | 681 | 21 (3.1) | 163 (23.9) | 497 (73.0) | 0.21 | 205 (15.1) | 1,157 (84.9) | 0.078 | 1.21 (0.98–1.51) | |
| Control | 699 | 14 (2.0) | 150 (21.5) | 535 (76.5) | 178 (12.7) | 1,220 (87.3) | 0.36 | |||
Note:
Hardy–Weinberg P-values in the control group.
Abbreviation: OR, odds ratio.
Demographic characteristics of the cognitive evaluation samples from the schizophrenia and control groups are summarized in Table S3. There were significant differences in terms of sex and years of education between the two groups (Ps<0.01). Consequently, we used these parameters as covariates to compare the RBANS scores between groups. Table S4 shows that patients had significantly poorer performance than controls on immediate memory, visuospatial skill, language, attention, delayed memory, and total RBANS score. We then examined whether rs1064395 plays an important role in cognitive dysfunction in patients with schizophrenia. ANCOVA was carried out with the rs1064395 genotypes (A/A + A/G vs G/G) as independent variables, the cognitive scores (RBANS total score and five index scores) as dependent variables, and age, gender, years of education, and duration of illness as covariates. Table 2 shows that patients with the A/A or A/G genotype of rs1064395 have lower scores of immediate memory, visuospatial skill, attention, and total RBANS score than those with the G/G genotype.
Table 2.
RBANS performance comparisons of rs1064395 genotypic groups in patients with schizophrenia
| RBANS | A/A + G/A (n=71) |
G/G (n=183) |
F-valuea | P-valueb | P-valuec |
|---|---|---|---|---|---|
| Total score | 291.45±40.11 | 320.42±30.91 | 46.03 | <0.01 | <0.01 |
| Immediate memory | 49.52±14.40 | 64.39±7.60 | 114.88 | <0.01 | <0.01 |
| Visuospatial skill | 56.58±8.85 | 59.49±5.66 | 12.10 | 0.001 | 0.006 |
| Language | 55.70±5.50 | 55.44±4.53 | 0.12 | 0.73 | |
| Attention | 64.37±18.99 | 73.19±18.09 | 14.52 | <0.01 | <0.01 |
| Delayed memory | 65.28±11.74 | 67.90±10.35 | 5.35 | 0.02 | 0.12 |
Notes: Data presented as .
F-values adjusted for age, gender, years of education, and duration of illness.
P-values not corrected for multiple testing.
P-values corrected after Bonferroni correction.
Abbreviation: RBANS, repeatable battery for the assessment of neuropsychological status.
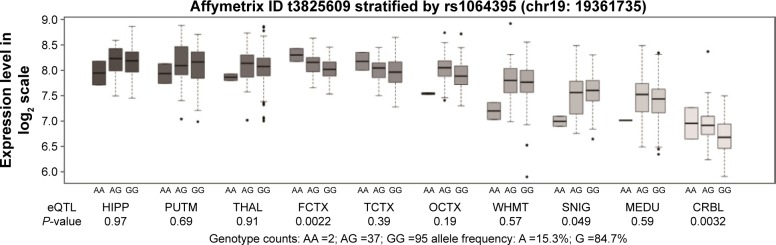
We then performed an eQTL analysis to investigate whether rs1064395 influences NCAN expression in the brain. As shown in Figure 1, we observed a significant association between rs1064395 and NCAN expression in the frontal cortex (P=0.0022, P=0.022 after Bonferroni correction) and cerebellar cortex (P=0.0032, P=0.032 after Bonferroni correction). Data show that carriers of the A allele have significantly higher levels of NCAN expression in the frontal and cerebellar cortex than those without the A allele.
Figure 1.
Association of rs1064395 with the NCAN mRNA expression level in ten brain regions (Affymetrix ID t3825609).
Abbreviations: CRBL, cerebellar cortex; FCTX, frontal cortex; HIPP, hippocampus; MEDU, the inferior olivary nucleus (sub-dissected from the medulla); OCTX, occipital cortex; PUTM, putamen (at the level of the anterior commissure); SNIG, substantia nigra; TCTX, temporal cortex; THAL, thalamus (at the level of the lateral geniculate nucleus); WHMT, intralobular white matter; Data were extracted from the BRAINEAC database (http://caprica.genetics.kcl.ac.uk/BRAINEAC/). Ramasamy A, Trabzuni D, Guelfi S, et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci. 2014;17(10):1418–1428.23
Discussion
Cognitive dysfunction has been widely reported to stably present in either patients with first-onset schizophrenia or those with chronic situation.26 Considerable evidence indicates that cognitive dysfunction is a core feature of schizophrenia, which can also serve as an endophenotype for the illness in genetic studies.27,28 In our previous work, we found that genes involved in neurodevelopment, such as brain-derived neurotrophic factor and ankyrin 3, have modulatory effects on executive function and working memory in patients with first onset schizophrenia.5,29 The gene and cognitive dysfunction associations strengthen the neurodevelopmental hypothesis of schizophrenia. A risk conferring allele A at locus rs1064395 within NCAN has been reported in the association with neurodevelopment, as well as the development of schizophrenia. The present study aimed to determine the role of the NCAN gene in shaping the developmental trajectories leading to schizophrenia.
NCAN rs1064395 has been reported to be a common risk factor for schizophrenia in Europeans.30,31 Here, we further confirmed rs1064395 as a risk locus for schizophrenia by utilizing a large-scale schizophrenia sample of European ancestry from PGC genome-wide association study. In the Han Chinese samples tested in this study, we failed to detect significance for rs1064395 in the susceptibility to schizophrenia. This suggests that rs1064395 is probably not a risk SNP for schizophrenia in Han Chinese. The A allele of rs1064395 in our control subjects was 12.7%, which is similar to the frequency in Han Chinese from the HapMap database (12.8%). We noticed that the frequency of the rs1064395 A allele in Europeans is reported to range from 14.7% to 15.2%.30,31 The inconsistent association of rs1064395 with schizophrenia between Chinese and European populations implies genetic heterogeneity of NCAN sequence variations across continental populations. As such, the population-specific factors, such as environmental exposure, diet, differences in genetic structure, population history, and culture, are likely to play an important role in the observed inconsistent replications.32
A recent neuroimaging study reported that NCAN risk status (carried with the A allele of rs1064395) is associated with higher folding in the right lateral occipital region and at a trend level for the left dorsolateral prefrontal cortex in patients with schizophrenia, but not in matched control subjects,9 implying that rs1064395 may be involved in cognitive disruption in schizophrenia. In this study, we found that schizophrenic patients with the A allele of NCAN rs1064395 have poorer cognitive processes, such as immediate memory, visuospatial skill, and attention, compared with those without the A allele. This result is in agreement with a recent study reporting that rs1064395A allele carriers exhibit poorer immediate and delayed verbal memory performance than those with the G/G genotype, implying that this SNP influences both neural processing and cognitive performance.33 We then carried out an eQTL analysis and found that rs1064395 is significantly associated with NCAN expression in the frontal and cerebellar cortex. Our findings suggest that NCAN rs1064395 may be a risk factor for the development of cognitive dysfunction in schizophrenia, whose regulatory effects are likely mediated via the frontal cortex and cerebellar cortex.
There is a large body of evidence from functional magnetic resonance imaging studies suggesting that cognitive deficits in patients with schizophrenia are associated with decreased activity of the frontal region, especially the dorsolateral prefrontal cortex, the ventrolateral prefrontal cortex, and the medial frontal cortex.34,35 In contrast, neuroimaging studies have shown that cortical cerebellar and global cerebellar volumes are significantly reduced in patients with schizophrenia compared to healthy controls.36 As the cerebellum represents at least 10% of the brain volume that contains 50% of its neurons,37 it has been established that cerebellar cortical volume reduction is implicated in cognitive dysfunction in schizophrenia.36 Moreover, the aberrant network connecting the frontal cortex and cerebellum is believed to be involved in the development of cognitive dysfunction,38 because cerebellar stimulation dynamically influences the medial frontal cortex in mice and clinically alleviates cognitive dysfunction in patients with schizophrenia.39,40 Both human and animal investigations suggest an important role for NCAN in the developing mammalian brain, and its expression is localized within the cognition-relevant brain regions, such as the cortex.41 The rs1064395 SNP is located within the 3′-UTR of NCAN. While the effect of the 3′-UTR on gene expression or function is not well known, it is possible that variants in this region could produce changes in messenger RNA processing.42,43 If accurate, rs1064395 may result in an alteration in NCAN expression, and by extension, account for the results we encountered in this study. However, no evidence supports the genuine effect of rs1064395 on NCAN, although some preliminary results indicate an effect on NCAN expression in the frontal cortex and cerebellum. Therefore, further research is required to verify this supposition.
When interpreting the results of this study, we would be remiss in not noting some limitations. First, the lack of a significant association between rs1064395 and schizophrenia may be caused by the modest sample size, possibly resulting in a type II error.44 Accordingly, our findings should be considered only preliminary and will require further investigations for validation in independent samples. Second, this case–control association study has the potential for population stratification, although all participants were of Han Chinese origin and from Eastern China. Therefore, we could not completely exclude the possibility of a population structure effect in our sample. Third, although we found that rs1064395 influences cognitive performance in patients with schizophrenia, the genuine effect of rs1064395 on NCAN is unclear. As such, the possibility that the observed effect was driven by another variation within NCAN that is in linkage disequilibrium with rs1064395 could not be fully excluded.45
Conclusion
Although we failed to replicate the findings of an association between rs1064395 and schizophrenia in Han Chinese. Our results indicate that this SNP may be a risk factor for cognitive dysfunction in patients with schizophrenia. While intriguing, these findings are still only suggestive, and further investigations are warranted to validate our findings to identify the precise mechanism by which rs1064395 influences cognitive performance in patients with schizophrenia.
Supplementary materials
Association of rs1064395 with schizophrenia in PGC database.
Abbreviations: GWAS, genome-wide association study; PGC, psychiatric genomics consortium; SCZ, schizophrenia.
Table S1.
Primers for the genotyping of rs1064395
| SNP ID | Primer | Primer sequences (5′>3′) |
|---|---|---|
| rs1064395 | PCRU | AGCCCAGTGCACATACCCAGTC |
| PCRL | GGGGAGGAAGGCAAGGTGAG | |
| SNP | t(gact)16 AACACTGAGCATCTCTCTACAATATGAC |
Notes: PCR amplification primers were marked by “PCRU” and “PCRL”, whereas SNP-specific oligonucleotide primer was marked by “SNP”. In the “(gact)n”, n means the number of “gact” repeats.
Abbreviations: PCR, polymerase chain reaction; SNP, single nucleotide polymorphism.
Table S2.
Logistic regression analysis of rs1064395 between schizophrenia and control groups
| SNP | Inheritance model | OR (95% CI) | P-value |
|---|---|---|---|
| rs1064395 | Codominant | ||
| G/A vs G/G | 0.85 (0.66–1.10) | 0.21 | |
| A/A vs G/G | 0.62 (0.31–1.23) | ||
| Dominant | |||
| G/A + A/A vs G/G | 0.83 (0.65–1.05) | 0.13 | |
| Recessive | |||
| A/A vs G/G + G/A | 0.64 (0.32–1.27) | 0.2 | |
| Log-additive | |||
| G/G vs G/A vs A/A | 0.83 (0.67–1.03) | 0.083 |
Note: Results adjusted by age and gender.
Abbreviations: CI, confidence interval; OR, odds ratio; SNP, single nucleotide polymorphism.
Table S3.
Demographic and clinical characteristics between schizophrenia and control groups
| Schizophrenia (n=254) | Controls (n=72) | Statistics | P-value | |
|---|---|---|---|---|
| Sex | 9.14 | <0.01 | ||
| Male | 118 | 40 | ||
| Female | 136 | 32 | ||
| Age (years)a | 34.0±8.7 | 26.2±5.6 | 1.86 | 0.18 |
| Years of educationa | 9.5±1.9 | 11.9±2.1 | −9.39 | <0.01 |
| Age at onset (years)a | 27.7±5.3 | |||
| Duration of illness (months)a | 45.8±16.5 |
Note:
Data presented as .
Table S4.
RBANS scores between schizophrenia and control groups
| RBANS | F-value | P-value | |
|---|---|---|---|
| Total score | 737.02 | <0.01 | |
| Cases | 312.32±36.09 | ||
| Controls | 463.69±21.92 | ||
| Immediate memory | 235.98 | <0.01 | |
| Cases | 60.24±11.99 | ||
| Controls | 89.21±8.59 | ||
| Visuospatial skill | 482.55 | <0.01 | |
| Cases | 58.68±6.82 | ||
| Controls | 88.74±11.51 | ||
| Language | 1,673.42 | <0.01 | |
| Cases | 55.52±4.81 | ||
| Controls | 93.63±8.28 | ||
| Attention | 84.45 | <0.01 | |
| Cases | 70.72±18.73 | ||
| Controls | 98.74±12.01 | ||
| Delayed memory | 234.13 | <0.01 | |
| Cases | 67.17±10.80 | ||
| Controls | 93.39±5.82 |
Note: Data presented as .
Abbreviation: RBANS, repeatable battery for the assessment of neuropsychological status.
Acknowledgments
We are deeply grateful to all participants. This work was supported by the National Natural Science Foundation of China (81471358), the Shanghai Science and Technology Commission Foundation (14411969000), the Shanghai Municipal Education Commission – Gaofeng Clinical Medicine Grant Support (20152530), the Shanghai Municipal Commission of Health and Family Planning Foundation (201540029), and the Shanghai Mental Health Center Foundation (2014-FX-03).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Bilder RM, Volavka J, Czobor P, et al. Neurocognitive correlates of the COMT Val(158)Met polymorphism in chronic schizophrenia. Biol Psychiatry. 2002;52(7):701–707. doi: 10.1016/s0006-3223(02)01416-6. [DOI] [PubMed] [Google Scholar]
- 2.Elvevag B, Goldberg TE. Cognitive impairment in schizophrenia is the core of the disorder. Crit Rev Neurobiol. 2000;14(1):1–21. [PubMed] [Google Scholar]
- 3.Prouteau A, Verdoux H, Briand C, et al. Cognitive predictors of psychosocial functioning outcome in schizophrenia: a follow-up study of subjects participating in a rehabilitation program. Schizophr Res. 2005;77(2–3):343–353. doi: 10.1016/j.schres.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Carter CS, Barch DM, CNTRICS Executive Committee Imaging biomarkers for treatment development for impaired cognition: report of the sixth CNTRICS meeting: Biomarkers recommended for further development. Schizophr Bull. 2012;38(1):26–33. doi: 10.1093/schbul/sbr109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu W, Zhang C, Yi Z, Li Z, Wu Z, Fang Y. Association between BDNF Val66Met polymorphism and cognitive performance in antipsychotic-naive patients with schizophrenia. J Mol Neurosci. 2012;47(3):505–510. doi: 10.1007/s12031-012-9750-4. [DOI] [PubMed] [Google Scholar]
- 6.Frischknecht R, Seidenbecher CI. The crosstalk of hyaluronan-based extracellular matrix and synapses. Neuron Glia Biol. 2008;4(3):249–257. doi: 10.1017/S1740925X09990226. [DOI] [PubMed] [Google Scholar]
- 7.Cubells JF, Sun X, Li W, et al. Linkage analysis of plasma dopamine beta-hydroxylase activity in families of patients with schizophrenia. Hum Genet. 2011;130(5):635–643. doi: 10.1007/s00439-011-0989-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muhleisen TW, Mattheisen M, Strohmaier J, et al. Association between schizophrenia and common variation in neurocan (NCAN), a genetic risk factor for bipolar disorder. Schizophr Res. 2012;138(1):69–73. doi: 10.1016/j.schres.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 9.Schultz CC, Muhleisen TW, Nenadic I, et al. Common variation in NCAN, a risk factor for bipolar disorder and schizophrenia, influences local cortical folding in schizophrenia. Psychol Med. 2014;44(4):811–820. doi: 10.1017/S0033291713001414. [DOI] [PubMed] [Google Scholar]
- 10.Cai J, Zhu Y, Zhang W, Wang Y, Zhang C. Comprehensive family therapy: an effective approach for cognitive rehabilitation in schizophrenia. Neuropsychiatr Dis Treat. 2015;11:1247–1253. doi: 10.2147/NDT.S83569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang C, Li Z, Shao Y, et al. Association study of tryptophan hydroxylase-2 gene in schizophrenia and its clinical features in Chinese Han population. J Mol Neurosci. 2011;43(3):406–411. doi: 10.1007/s12031-010-9458-2. [DOI] [PubMed] [Google Scholar]
- 12.Randolph C, Tierney MC, Mohr E, Chase TN. The repeatable battery for the assessment of neuropsychological status (RBANS): preliminary clinical validity. J Clin Exp Neuropsychol. 1998;20(3):310–319. doi: 10.1076/jcen.20.3.310.823. [DOI] [PubMed] [Google Scholar]
- 13.Cheng Y, Wu W, Wang J, Feng W, Wu X, Li C. Reliability and validity of the Repeatable Battery for the Assessment of Neuropsychological Status in community-dwelling elderly. Arch Med Sci. 2011;7(5):850–857. doi: 10.5114/aoms.2011.25561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang XY, Chen da C, Tan YL, et al. BDNF Polymorphisms are associated with cognitive performance in schizophrenia patients versus healthy controls. J Clin Psychiatry. 2016;77(8):e1011–e1018. doi: 10.4088/JCP.15m10269. [DOI] [PubMed] [Google Scholar]
- 15.Hui L, Han M, Huang XF, et al. Association between DβH 5′-insertion/deletion polymorphism and cognition in patients with chronic schizophrenia. J Clin Psychiatry. 2016;77(3):379–385. doi: 10.4088/JCP.14m09629. [DOI] [PubMed] [Google Scholar]
- 16.Zhang XY, Chen DC, Xiu MH, et al. Cognitive function, plasma MnSOD activity, and MnSOD Ala-9Val polymorphism in patients with schizophrenia and normal controls. Schizophr Bull. 2014;40(3):592–601. doi: 10.1093/schbul/sbt045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang C, Wu Z, Zhao G, Wang F, Fang Y. Identification of IL6 as a susceptibility gene for major depressive disorder. Sci Rep. 2016;6:31264. doi: 10.1038/srep31264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang C, Wang Z, Hong W, Wu Z, Peng D, Fang Y. ZNF804A Genetic Variation Confers Risk to Bipolar Disorder. Mol Neurobiol. 2016;53(5):2936–2943. doi: 10.1007/s12035-015-9193-3. [DOI] [PubMed] [Google Scholar]
- 19.Schizophrenia psychiatric genome-wide association study (GWAS) Consortium Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43(10):969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fornito A, Bullmore ET. Reconciling abnormalities of brain network structure and function in schizophrenia. Curr Opin Neurobiol. 2015;30:44–50. doi: 10.1016/j.conb.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 21.Zhang C, Lu W, Wang Z, et al. A comprehensive analysis of NDST3 for schizophrenia and bipolar disorder in Han Chinese. Transl Psychiatry. 2016;6:e701. doi: 10.1038/tp.2015.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang C, Zhang DF, Wu ZG, et al. Complement factor H and susceptibility to major depressive disorder in Han Chinese. Br J Psychiatry. 2016;208(5):446–452. doi: 10.1192/bjp.bp.115.163790. [DOI] [PubMed] [Google Scholar]
- 23.Ramasamy A, Trabzuni D, Guelfi S, et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci. 2014;17(10):1418–1428. doi: 10.1038/nn.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15(2):97–98. doi: 10.1038/sj.cr.7290272. [DOI] [PubMed] [Google Scholar]
- 25.Sole X, Guino E, Valls J, Iniesta R, Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22(15):1928–1929. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- 26.Kurtz MM. Neurocognitive impairment across the lifespan in schizophrenia: an update. Schizophr Res. 2005;74(1):15–26. doi: 10.1016/j.schres.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 27.Braff DL, Freedman R, Schork NJ, Gottesman II. Deconstructing schizophrenia: an overview of the use of endophenotypes in order to understand a complex disorder. Schizophr Bull. 2007;33(1):21–32. doi: 10.1093/schbul/sbl049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burdick KE, Goldberg JF, Harrow M, Faull RN, Malhotra AK. Neurocognition as a stable endophenotype in bipolar disorder and schizophrenia. J Nerv Ment Dis. 2006;194(4):255–260. doi: 10.1097/01.nmd.0000207360.70337.7e. [DOI] [PubMed] [Google Scholar]
- 29.Zhang C, Cai J, Zhang J, et al. Genetic modulation of working memory deficits by ankyrin 3 gene in schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry. 2014;50:110–115. doi: 10.1016/j.pnpbp.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 30.Rietschel M, Mattheisen M, Degenhardt F, et al. Association between genetic variation in a region on chromosome 11 and schizophrenia in large samples from Europe. Mol Psychiatry. 2012;17(9):906–917. doi: 10.1038/mp.2011.80. [DOI] [PubMed] [Google Scholar]
- 31.Shi J, Levinson DF, Duan J, et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009;460(7256):753–757. doi: 10.1038/nature08192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li M, Luo XJ, Xiao X, et al. Allelic differences between Han Chinese and Europeans for functional variants in ZNF804A and their association with schizophrenia. Am J Psychiatry. 2011;168(12):1318–1325. doi: 10.1176/appi.ajp.2011.11030381. [DOI] [PubMed] [Google Scholar]
- 33.Raum H, Dietsche B, Nagels A, et al. A genome-wide supported psychiatric risk variant in NCAN influences brain function and cognitive performance in healthy subjects. Hum Brain Mapp. 2015;36(1):378–390. doi: 10.1002/hbm.22635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buchy L, Hawco C, Joober R, Malla A, Lepage M. Cognitive insight in first-episode schizophrenia: further evidence for a role of the ventrolateral prefrontal cortex. Schizophr Res. 2015;166(1–3):65–68. doi: 10.1016/j.schres.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 35.Li X, Xiao YH, Zhao Q, Leung AW, Cheung EF, Chan RC. The neuroplastic effect of working memory training in healthy volunteers and patients with schizophrenia: Implications for cognitive rehabilitation. Neuropsychologia. 2015;75:149–162. doi: 10.1016/j.neuropsychologia.2015.05.029. [DOI] [PubMed] [Google Scholar]
- 36.Laidi C, d’Albis MA, Wessa M, et al. Cerebellar volume in schizophrenia and bipolar I disorder with and without psychotic features. Acta Psychiatr Scand. 2015;131(3):223–233. doi: 10.1111/acps.12363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li WK, Hausknecht MJ, Stone P, Mauk MD. Using a million cell simulation of the cerebellum: network scaling and task generality. Neural Netw. 2013;47:95–102. doi: 10.1016/j.neunet.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parker KL. Timing Tasks Synchronize Cerebellar and Frontal Ramping Activity and Theta Oscillations: Implications for Cerebellar Stimulation in Diseases of Impaired Cognition. Front Psychiatry. 2015;6:190. doi: 10.3389/fpsyt.2015.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rogers TD, Dickson PE, McKimm E, et al. Reorganization of circuits underlying cerebellar modulation of prefrontal cortical dopamine in mouse models of autism spectrum disorder. Cerebellum. 2013;12(4):547–556. doi: 10.1007/s12311-013-0462-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Demirtas-Tatlidede A, Freitas C, Cromer JR, et al. Safety and proof of principle study of cerebellar vermal theta burst stimulation in refractory schizophrenia. Schizophr Res. 2010;124(1–3):91–100. doi: 10.1016/j.schres.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cichon S, Muhleisen TW, Degenhardt FA, et al. Genome-wide association study identifies genetic variation in neurocan as a susceptibility factor for bipolar disorder. Am J Hum Genet. 2011;88(3):372–381. doi: 10.1016/j.ajhg.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Conne B, Stutz A, Vassalli JD. The 3′ untranslated region of messenger RNA: A molecular ‘hotspot’ for pathology? Nat Med. 2000;6(6):637–641. doi: 10.1038/76211. [DOI] [PubMed] [Google Scholar]
- 43.Zhu Y, Wang Z, Ni J, et al. Genetic variant in NDUFS1 gene is associated with schizophrenia and negative symptoms in Han Chinese. J Hum Genet. 2015;60(1):11–16. doi: 10.1038/jhg.2014.94. [DOI] [PubMed] [Google Scholar]
- 44.Ni J, Hu S, Zhang J, Tang W, Lu W, Zhang C. A Preliminary Genetic Analysis of Complement 3 Gene and Schizophrenia. PLoS One. 2015;10(8):e0136372. doi: 10.1371/journal.pone.0136372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dannlowski U, Kugel H, Grotegerd D, et al. NCAN cross-disorder risk variant is associated with limbic gray matter deficits in healthy subjects and major depression. Neuropsychopharmacology. 2015;40(11):2510–2516. doi: 10.1038/npp.2015.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Association of rs1064395 with schizophrenia in PGC database.
Abbreviations: GWAS, genome-wide association study; PGC, psychiatric genomics consortium; SCZ, schizophrenia.
Table S1.
Primers for the genotyping of rs1064395
| SNP ID | Primer | Primer sequences (5′>3′) |
|---|---|---|
| rs1064395 | PCRU | AGCCCAGTGCACATACCCAGTC |
| PCRL | GGGGAGGAAGGCAAGGTGAG | |
| SNP | t(gact)16 AACACTGAGCATCTCTCTACAATATGAC |
Notes: PCR amplification primers were marked by “PCRU” and “PCRL”, whereas SNP-specific oligonucleotide primer was marked by “SNP”. In the “(gact)n”, n means the number of “gact” repeats.
Abbreviations: PCR, polymerase chain reaction; SNP, single nucleotide polymorphism.
Table S2.
Logistic regression analysis of rs1064395 between schizophrenia and control groups
| SNP | Inheritance model | OR (95% CI) | P-value |
|---|---|---|---|
| rs1064395 | Codominant | ||
| G/A vs G/G | 0.85 (0.66–1.10) | 0.21 | |
| A/A vs G/G | 0.62 (0.31–1.23) | ||
| Dominant | |||
| G/A + A/A vs G/G | 0.83 (0.65–1.05) | 0.13 | |
| Recessive | |||
| A/A vs G/G + G/A | 0.64 (0.32–1.27) | 0.2 | |
| Log-additive | |||
| G/G vs G/A vs A/A | 0.83 (0.67–1.03) | 0.083 |
Note: Results adjusted by age and gender.
Abbreviations: CI, confidence interval; OR, odds ratio; SNP, single nucleotide polymorphism.
Table S3.
Demographic and clinical characteristics between schizophrenia and control groups
| Schizophrenia (n=254) | Controls (n=72) | Statistics | P-value | |
|---|---|---|---|---|
| Sex | 9.14 | <0.01 | ||
| Male | 118 | 40 | ||
| Female | 136 | 32 | ||
| Age (years)a | 34.0±8.7 | 26.2±5.6 | 1.86 | 0.18 |
| Years of educationa | 9.5±1.9 | 11.9±2.1 | −9.39 | <0.01 |
| Age at onset (years)a | 27.7±5.3 | |||
| Duration of illness (months)a | 45.8±16.5 |
Note:
Data presented as .
Table S4.
RBANS scores between schizophrenia and control groups
| RBANS | F-value | P-value | |
|---|---|---|---|
| Total score | 737.02 | <0.01 | |
| Cases | 312.32±36.09 | ||
| Controls | 463.69±21.92 | ||
| Immediate memory | 235.98 | <0.01 | |
| Cases | 60.24±11.99 | ||
| Controls | 89.21±8.59 | ||
| Visuospatial skill | 482.55 | <0.01 | |
| Cases | 58.68±6.82 | ||
| Controls | 88.74±11.51 | ||
| Language | 1,673.42 | <0.01 | |
| Cases | 55.52±4.81 | ||
| Controls | 93.63±8.28 | ||
| Attention | 84.45 | <0.01 | |
| Cases | 70.72±18.73 | ||
| Controls | 98.74±12.01 | ||
| Delayed memory | 234.13 | <0.01 | |
| Cases | 67.17±10.80 | ||
| Controls | 93.39±5.82 |
Note: Data presented as .
Abbreviation: RBANS, repeatable battery for the assessment of neuropsychological status.