Abstract
Purpose
Progression rate of age-related macular degeneration (AMD) varies substantially, yet its association with genetic variation has not been widely examined.
Methods
We tested whether progression rate from intermediate AMD to geographic atrophy (GA) or choroidal neovascularization (CNV) was correlated with genotype at seven single nucleotide polymorphisms (SNPs) in the four genes most strongly associated with risk of advanced AMD. Cox proportional hazards survival models examined the association between progression time and SNP genotype while adjusting for age and sex and accounting for variable follow-up time, right censored data, and repeated measures (left and right eyes).
Results
Progression rate varied with the number of risk alleles at the CFH:rs10737680 but not the CFH:rs1061170 (Y402H) SNP; individuals with two risk alleles progressed faster than those with one allele (hazard ratio [HR] = 1.61, 95% confidence interval [CI] = 1.08–2.40, P < 0.02, n = 547 eyes), although this was not significant after Bonferroni correction. This signal was likely driven by an association at the correlated protective variant, CFH:rs6677604, which tags the CFHR1-3 deletion; individuals with at least one protective allele progressed more slowly. Considering GA and CNV separately showed that the effect of CFH:rs10737680 was stronger for progression to CNV.
Conclusions
Results support previous findings that AMD progression rate is influenced by CFH, and suggest that variants within CFH may have different effects on risk versus progression. However, since CFH:rs10737680 was not significant after Bonferroni correction and explained only a relatively small portion of variation in progression rate beyond that explained by age, we suggest that additional factors contribute to progression.
Keywords: age-related macular degeneration (AMD), genetics, progression
Age-related macular degeneration (AMD) is a major cause of visual loss in older adults.1 This complex neurodegenerative eye disease encompasses a wide range of phenotypic traits, although stages of the disease are often broadly defined as early, intermediate, and advanced AMD.2 Progression through early and intermediate stages is generally characterized by expansion in size and number of extracellular deposits (drusen) and changes to pigmentation of the retinal pigment epithelium (RPE), until in advanced stages there is either extensive RPE cell atrophy in nonneovascular AMD (geographic atrophy, GA) and/or abnormal blood vessel growth under the retina in neovascular AMD (choroidal neovascularization, CNV).3 Despite substantial phenotypic heterogeneity, most studies of the genetic epidemiology of AMD have focused on predicting risk of advanced disease by comparing advanced cases with controls, rather than risk of early or intermediate stages of the disease or disease progression.2
Advanced AMD is highly heritable, with an estimated 70% of variance in disease risk attributed to genetic variation.4 Accordingly, numerous genetic variants associated with risk of advanced AMD have been identified, including common single nucleotide variants (SNVs) of large effect in the complement factor H (CFH),5–9 complement component 2/factor B (C2/CFB),10,11 complement component 3 (C3),12,13 and age-related maculopathy susceptibility 2 (ARMS2)/HtrA serine peptidase 1 (HTRA1) regions.14–16 The International AMD Genetics Consortium identified 19 independently associated common risk variants by genome-wide meta-analysis of >70,000 cases and controls,17 and most recently, a total of 52 variants associated with risk of advanced AMD in an exome-chip analysis of common and rare variation.18 Demographic and environmental risk factors for advanced AMD have also been identified, including increased age, smoking, body mass index (BMI), and diet.19,20
Known risk factors predict the presence or absence of advanced AMD in cross-sectional cohorts of cases and controls relatively well compared to other complex diseases.2,18 Whether the same or additional genetic factors also predict progression from early-/intermediate- to late-stage disease has not been extensively tested. However, rates of disease progression are highly variable; while some individuals progress quickly to late-stage AMD, others progress slowly over many years, or never progress to advanced disease21 at all. Establishing the genetic basis of progression to advanced disease will increase our understanding of the underlying disease process and potentially identify novel targets for drug design. Understanding of the genetic etiology of AMD progression will also allow individuals at risk of rapid progression to be identified, enabling monitoring and treatment strategies to be improved.
Previous studies found that progression of AMD grade was associated with variants in the CFH,22–29 ARMS2/HTRA1,22–24,26,28,30 C2/CFB,23,24,26,28,31 C3,23,24,26,28,31 APOE,32 LIPC and ABCA1,26 and COL8A1 and RAD51B genes.33 Advanced age, smoking history, education level, BMI, and antioxidant use have also been associated with AMD progression.23,24,28,34,35 However, only some of these studies considered time to progression22–26,28,33 rather than simply testing whether or not progression occurred over a single fixed time period. Furthermore, many of these studies used the AREDS dataset22–24,26,28,31,33; replication of previous findings in additional cohorts with time-to-event data has been limited. Additional replication is therefore necessary.
We used longitudinal data to test for variation in progression rate from intermediate to advanced AMD and genotype at seven variants in the four genes most strongly associated with risk of advanced AMD,18 accounting for age, sex, variable follow-up time, and variation between left and right eyes within individuals. We thereby tested whether previous findings were replicated in an independent cohort.
Methods
Participants and Phenotype
Subjects of European ancestry were recruited from the Vanderbilt Eye Institute (VEI) and the University of Miami Bascom Palmer Eye Institute (BPEI). Consent was obtained from all participants, all procedures conformed to the Declaration of Helsinki, and research was carried out under protocols approved by the Institutional Review Boards at each institution. All participants were examined by a retina specialist using slit-lamp biomicroscopy and dilated fundus examination. Fundus images were graded using the modified Age-Related Eye Disease Study (AREDS) scale (Clinical Age-Related Maculopathy Scaling, CARMS) where grade 1 corresponds to unaffected controls, grade 2 to early AMD, grade 3 to intermediate AMD, grade 4 to GA, and grade 5 to CNV.3,20,36 Patients were reexamined approximately every 6 to 12 months based on standard clinical care. Data were also collected retrospectively to expand the time span of monitored cases. Follow-up time ranged from 1 month to 13 years. Progression of an eye was defined as a change from grade 3 (intermediate AMD) to either grade 4 (GA) or 5 (CNV), and progression time as the number of months from the first grade 3 exam to the first grade 4 or grade 5 exam. Nonprogressing eyes were defined as those that remained a grade 3 by the most recent exam. A total of 577 eyes (392 individuals) met criteria for inclusion in our progression study (at least one grade 3 exam and at least one follow-up exam).
Variant Selection
Seven variants associated with risk of advanced AMD were evaluated for their effect on progression rate (Table 1). We tested the four common SNVs that were most strongly associated with risk of advanced AMD (largest odds ratios) in a large meta-analysis of >17,000 cases and >60,000 controls17,18: rs10490924 in the ARMS2/HTRA1 region of chromosome 10, rs10737680 in the CFH region on chromosome 1, rs429608 in C2/CFB/SKIV2L on chromosome 6, and rs2230199 in C3 on chromosome 19. We also tested the three additional variants in these regions that were tested for effects on AMD progression previously,24,25,30,31 thereby allowing comparison between studies: the independently associated nonsynonymous CFH:rs1061170 (Y402H) polymorphism and two protective variants in the C2-CFB region (C2:rs9332739 and CFB:rs641153), which are usually considered to be the primary causal variants associated with risk of advanced AMD in these regions.7,10,11
Table 1.
Details of the Seven AMD Risk Variants Tested for Association With Progression Rate From Intermediate to Advanced AMD
DNA Extraction and Genotyping
A total of 392 individuals were genotyped at ARMS2:rs10490924, CFH:rs10737680, C2:rs429608, C2:rs9332739, and C3:rs2230199 by at least one of three methods: (1) a 19 SNV Sequenom MassARRAY (Sequenom, Inc., San Diego, CA, USA) (n = 210 individuals); (2) HumanCoreExome chip genotyping (Illumina, San Diego, CA, USA) (n = 257 samples genotyped using a custom-modified Illumina HumanCoreExome chip as part of 50,000 individuals genotyped at the Center for Inherited Disease Research (CIDR) by the International AMD Gene Consortium, IAMDGC,18 and n = 127 samples genotyped at the University of Miami); and (3) Taqman (Applied Biosystems, Foster City, CA, USA) (n = 32). All SNVs had call rates > 98%. Genotypes at the CFH:rs1061170 and CFB:rs641153 single nucleotide polymorphisms (SNPs) were imputed from the exome-chip data using the 1000 Genomes reference panel,37 SHAPEIT for phasing,38 and IMPUTE2 for imputation39; both variants were imputed with info quality scores > 0.99.
Samples genotyped on multiple platforms were checked for concordance and excluded if mismatches occurred at any SNP. Analyses were restricted to unrelated individuals of (self-reported) European ancestry since the allele frequency of genetic risk variants varies with ethnicity. Ethnicity of samples with exome-chip data was confirmed using Eigenstrat40; any samples that did not cluster with individuals of Northern European ancestry from the 1000 Genomes database (CEU population) were excluded. In total, 20 individuals were excluded from analyses, leaving a final sample of 372 genotyped individuals (547 eyes); 14 individuals did not cluster with the CEU population, 2 showed mismatches between genotyping platforms, and 4 showed a mismatch between X-chromosome homozygosity in PLINK41 and recorded sex.
Statistical Analysis
Cox proportional hazards regression models were used to test whether progression rate from intermediate to advanced AMD varied with genotype at each of seven SNPs. Models were fit using the coxph function in the R-package Survival, version 2.38.42 Cox proportional hazards models assess the time it takes for an event (here, progression) to occur, and whether this is associated with covariates. The response variable was coded as a two-column vector of “survival” time that consisted of (a) follow-up time in months from the first grade 3 exam to either the exam at which progression was recorded, or the most recent exam, and (b) whether or not an eye had progressed by that time. The use of Cox proportional hazards models allowed both variable follow-up time and right censoring of data in instances in which an eye had not progressed by the most recent exam to be accounted for. Any correlation in progression rate across contralateral eyes43 was taken into account using the generalized estimating equations (GEE) function available for the Cox proportional hazards model by including a cluster (repeated measures) term for individual ID that averaged the response (progression rate) across an individual's two eyes.44 Hazard ratios (HR) (and 95% confidence intervals [CI]) are reported for each predictor and measure the ratio of progression times per unit change. Genotype at each SNP was coded as a three-level factor corresponding to zero, one, or two risk alleles (homozygous reference, heterozygous or homozygous for the risk allele). For imputed variants, all Y402H genotypes and the majority of the rs641153 genotypes (except for three) were imputed with 100% probability, and were therefore also treated as three-level factors in analyses (the three genotypes in dosage format were hard-called as the most likely genotype). For SNPs for which the sample size of individuals with a particular genotype was less than 5, we pooled genotype groups to create two-level factors; for C2:rs429608, C2:rs9332739, and CFB:rs641153 we therefore we pooled individuals with zero risk alleles with those that carried one risk allele (Table 1). For three-level factors, the model reference level was initially set to zero risk alleles, allowing the contrast in progression between subjects with one or two versus no risk alleles to be estimated. Models were then releveled to set individuals with one risk allele as the reference level to allow contrasts between individuals with two versus one risk allele to be estimated. We also tested an additive disease model by treating genotype as a covariate; however, results were qualitatively similar (data not presented). Models controlled for variation in progression rate due to age (at first grade 3 exam) and sex by including these variables as main effects. Initially, we also adjusted models for ascertainment site (Miami or Vanderbilt). However, progression rate did not vary between sites, and results were quantitatively similar with and without this term in the model; we therefore excluded this term from the final models presented here. We confirmed that data met the proportional hazards assumption by testing the correlation between model residuals and time. Hazard ratios and 95% CI were calculated. Survival curves of the predicted progression rate for individuals of each genotype were plotted using the survfit function in the Survival package. Initially, separate models were run for each SNV (including both age and sex), followed by a model including all seven variants. We also ran separate models for progression to GA and to CNV only where progression to GA analyses included eyes that had not progressed by the most recent exam and those that progressed from grade 3 to grade 4, while progression to CNV analyses included eyes that had not progressed by the most recent exam and eyes that progressed directly from grade 3 to grade 5. To estimate the predictive power of our final model we calculated area under the receiver operating characteristic (ROC) curves (AUC) at yearly follow-up time points (1–5 years) using the R-package survivalROC45 and compared this to the AUC for models without genotype (including age and sex only). We conducted a power analysis using the R-package survSNP.46 All analyses were run in R version 3.0.1, available at https://www.r-project.org/.47
Results
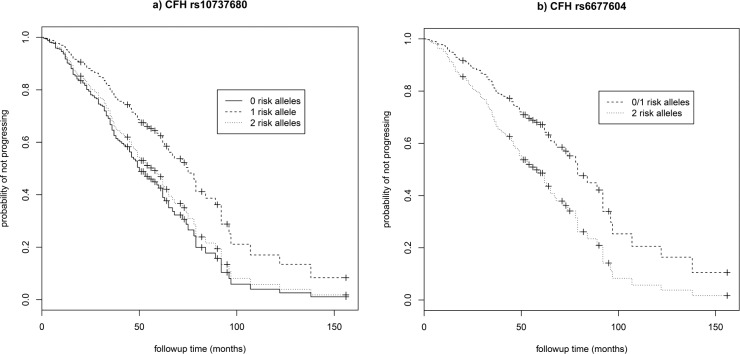
A total of 547 eyes (255 right eyes and 292 left eyes) from 372 individuals (159 males and 213 females) were available for analysis, of which 167 eyes from 137 individuals progressed over the study period (72 to grade 4 and 95 to grade 5) with a mean progression time of 32.2 (±25.2) months (range, 1–138 months). Age at first grade 3 exam ranged from 53 to 95 years (mean age per eye = 76.3 ± 7.6). Individuals with two risk alleles at CFH:rs10737680 progressed at a significantly faster rate than those with one allele (HR = 1.61, 95% CI = 1.08–2.40, P = 0.02; Table 2; Fig. 1a), although this was not significant after Bonferroni correction for multiple tests (n = 7). There was no significant difference in progression rate between individuals with no CFH:rs10737680 risk alleles and either one or two risk alleles, but the sample size of individuals with no CFH:rs10737680 risk alleles was small (n = 31 eyes/20 individuals; Tables 1, 2; Supplementary Table S1). Concordance of the Cox proportional hazards model including CFH, sex, and age was 0.65 (±SE 0.03). Time-dependent ROC curves estimated that the AUC at 3 years (mean follow-up time in our dataset) for a model including age, sex, and CFH:rs10737680 was slightly higher (0.67; AUC range, 0.67–0.71 for 1–5 year follow-up time) compared to a model accounting for age and sex only (AUC = 0.64; range, 0.64–0.67; Fig. 2). Since the CFH:rs10737680 variant is correlated with two distinct protective haplotypes, CFH:p.I62V that is tagged by CFH:rs800292, and the CFHR1-3 deletion tagged by CFH:rs6677604, we further evaluated the signal at this position by testing progression rate with respect to these two (uncorrelated) SNPs that were available in our imputed dataset. Individuals with two risk alleles (no protective alleles) at CFH:rs6677604 (n = 288) progressed significantly faster than those with fewer than two risk alleles (one or two protective alleles, n = 81; HR = 1.81, 95% CI = 1.11–2.94, P = 0.02; Fig. 1b) where individuals with one or two protective alleles were pooled due to small sample size (n = 5 and 76, respectively). There was no association between progression rate and genotype at the rs800292 SNP (HR = 0.95, 95% CI = 0.61–1.47, P = 0.81).
Table 2.
Regression Coefficients and Hazard Ratios (HR) From Cox Proportional Hazards Models Testing Whether Progression Rate From Intermediate to Advanced (Grade 4 or 5) Varied With Genotype at 7 Variants Associated With Risk of Advanced AMD After Controlling for Variation Due to Age and Sex
Figure 1.
Survival curves for progression to advanced AMD (GA or CNV) and genotype at two associated variants in the CFH locus. Survival curves showing the predicted progression rate from intermediate (grade 3) to advanced (grade 4 or 5) AMD with respect to the number of risk alleles at (a) nominally significant CFH:rs10737680 (P = 0.02, unadjusted for multiple testing), and (b) the associated SNP CFH:rs6677604 (P = 0.02), for females 67 years of age.
Figure 2.
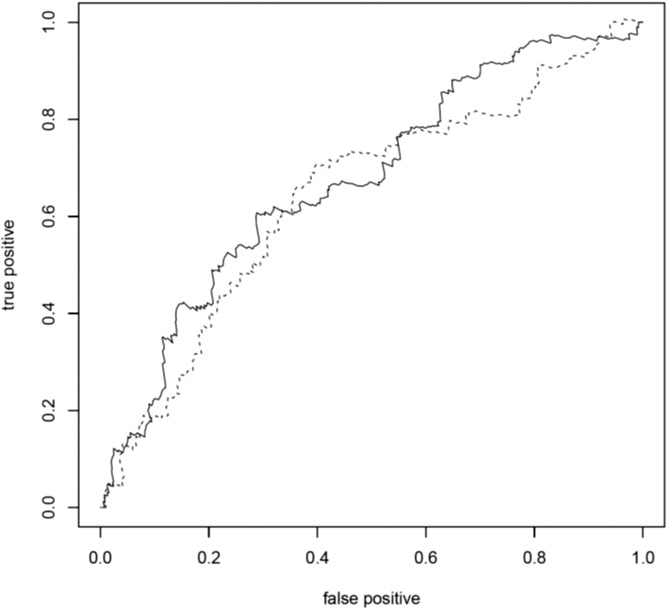
Receiver operating characteristic curve. Receiver operating characteristic curves of the true-positive rate (sensitivity) versus false-positive rate (1 − specificity) showing the ability of a model including age, sex, and number of CFH:rs10737680 risk alleles (solid line), and a model including age and sex only (dashed line), to distinguish between eyes that progressed to advanced AMD and those that did not progress at 3-year follow-up. The closer to the left and top borders the line is, the better the model performs in distinguishing between progressors and nonprogressors. Areas under the curve were 0.67 (age, sex, and CFH:rs10737680 genotype) and 0.64 (age and sex only), respectively.
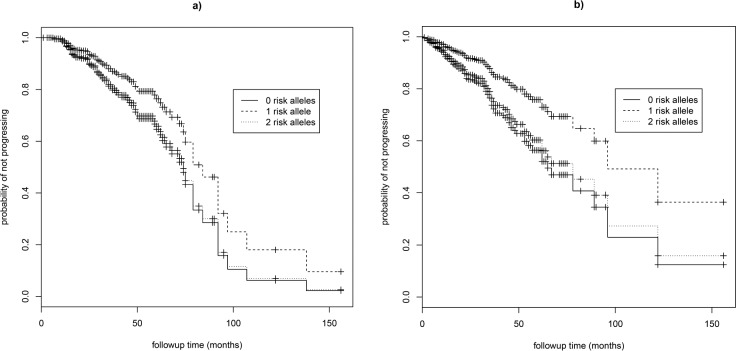
The number of risk alleles at the ARMS2:rs10490924, CFH:rs1061170, C2:rs429608, C2:rs9332739, CFB:rs641153, and C3:rs2230199 SNPs did not significantly predict progression rate in our dataset (Table 2; Supplementary Fig. S1). Older individuals had significantly higher HR and therefore progressed at a faster rate (HR = 1.05–1.06 corresponding to a 1-year change in age, 95% CI = 1.03–1.08, P < 0.001; Table 2). Progression rate did not vary with sex (Table 2). A model including all seven variants produced quantitatively similar results to models run for each variant separately (Supplementary Table S2). When separate models were run for progression to GA (grade 4) and progression to CNV (grade 5), the number of risk alleles at CFH:rs10737680 was significant only for predicting progression rate to CNV (two versus one allele: HR = 1.83, 95% CI = 1.09–3.05, P = 0.02; Table 3; Fig. 3; n = 452 eyes), not for progression to GA, although the effect was in the same direction (HR = 1.56, 95% CI = 0.85–2.85, P = 0.15; Table 3; n = 475 eyes). Hazard ratios for the other six variants were similar for both models of progression to GA and to CNV (Table 3). When separate GA and CNV models were run for CFH:rs6677604, results were similar to those for CFH:rs10737680. For progression to CNV, individuals with two risk alleles (no protective alleles) progressed faster than those with one or more protective alleles (HR = 2.46, 95% CI = 1.22–4.93, P = 0.01), while for progression to GA there was no significant association with genotype, although results tended to be in the same direction (HR = 1.49, 95% CI = 0.76–2.90, P = 0.24). Similar results were found when we tested the effect of CFH on progression to CNV by excluding grade 5 eyes that showed concomitant GA (n = 7; results not presented).
Table 3.
Regression Coefficients and Hazard Ratios (HR) From Cox Proportional Hazards Models Testing Whether Progression Rate From Intermediate (Grade 3) to Advanced Dry AMD (Grade 4) or Advanced Wet AMD (Grade 5) Varied With Genotype at 7 Variants Associated With Risk of Advanced AMD
Figure 3.
Survival curves for GA and CNV. Survival curves showing the progression rate from intermediate (grade 3) to (a) GA (grade 4) AMD and (b) CNV (grade 5) AMD with respect to the number of risk alleles at CFH:rs10737680 for females of mean age (67 years of age).
Power analyses estimated that we had at least 80% power to detect HR from 1.5 to 2 given a range of risk allele frequencies (0.3–0.6 and 0.1–0.8, respectively) assuming an event error rate of 0.31, sample size of 372 individuals, and median time to progression of 25 months.
Discussion
We tested whether progression rate from intermediate (nonneovascular) AMD to advanced AMD (GA or CNV) varied with genotype at seven variants in four gene regions strongly associated with risk of advanced AMD. We demonstrate that, after adjusting for age, individuals with two copies of the CFH:rs10737680 risk allele progressed faster than those with one copy. Although this association was of moderate effect size (HR = 1.6) and remained when genotypes at the other six SNPs were included in the model, the term was not significant after Bonferroni correction for multiple tests. Furthermore, the inclusion of CFH genotype did not substantially increase the predictive power of a model (AUC) that already included age at exam. However, despite nominal significance, these results do replicate an association between progression and the CFH:rs10737680 SNP in published work on the AREDS dataset (where AMD grades were reclassified using the CARMS grading system) in which progression rate from early/intermediate to advanced AMD was higher for individuals with one or two copies of the CFH:rs1410996 variant (a 100% proxy for rs10737680) than for those with no copies of the risk allele.24,33 By contrast, in our study, individuals with no rs10737680 risk alleles had a progression rate similar to those with two risk alleles. However, because the CFH risk allele occurs at high allele frequency, especially in a cohort of individuals with AMD, our sample of individuals with no copies of the CFH risk allele was relatively small (20 individuals), potentially preventing a robust estimate of progression for this group.
The CFH region consists of many highly correlated variants in linkage disequilibrium, and signals can be challenging to separate.48,49 However, secondary analyses suggested that association between progression rate and CFH:rs10737680 was driven primarily by an effect of the deletion CFHR3/CFHR1 tagged by CFH:rs6677604. This SNP has a protective effect on AMD risk, where the minor allele (A) increases the expression of CFH levels, protecting against risk of developing AMD.50,51 Our analyses suggested that this variant also has an impact on time to progression since individuals with two risk alleles (no protective alleles) progressed faster than those with fewer than two risk alleles (one or two protective alleles).
In contrast to CFH:rs10737680, we did not find an association between progression rate and the CFH:rs1061170 (Y402H) variant. Y402H is strongly associated with risk of advanced AMD5–7 and has also been associated with the probability of progression in other studies,22,23,29 with the exception of Farwick et al.30 Y402H was also correlated with progression rate in time-varying models of the AREDS24,26,28 and Beaver Dam datasets.25 However, in studies that considered the effect of both CFH variants, rs10737680 tended to have a stronger effect than Y402H on both the probability of progression to advanced AMD and progression rate, especially in multivariate models.24,28,29,33,52 Therefore, although the two SNPs are in linkage disequilibrium (r2 = 0.41/D′ = 1 in Northern European samples from the 1000 Genomes dataset), rs10737680 appears to be the CFH variant most strongly associated with progression of AMD through its correlation with CFH:rs667604.
We did not find any evidence that progression rate from intermediate to advanced AMD varied with genotype at variants in three other major genes associated with risk of advanced AMD (ARMS2, C2/CFB, and C3), despite having 80% power to detect a moderate effect of common variants (HR = 1.5–2). We were, however, underpowered to detect a moderate effect size for rare variants such as C2:rs9332739 and C2:rs641153. By contrast, in the AREDS dataset, progression rate was higher for individuals with at least one copy of the ARMS2:rs10490924 risk allele and for those with two versus no copies of the C3:rs2230199 risk allele, while individuals with at least one copy of either of the protective variants in the C2/CFB region (rs9332739 and rs641153) progressed more slowly.24,26,28,33 The probability of progression to late AMD also varied with genotype at ARMS2:rs10490924 but not C3:rs2230199 in the MARS (Münster Ageing and Retina Study) cohort,30 and with genotype at ARMS2:rs1049092, C3:rs2230199, and C2:rs9332739 in the AREDS data.23,31 Variation in findings between studies may reflect differing sample size and hence power, additional covariates, and/or differing analytical methods. Here we tested time to progression using survival models, rather than the probability of progression over a fixed time period. We also included both left and right eyes for each individual rather than including only the more severely affected eye, allowing us to average the progression rate of eyes within individuals, thereby accounting for individual-level variation. Additionally, we included only eyes that were grade 3 at baseline to minimize variation in the dataset, whereas some studies also included eyes that were grade 1 or 2 at baseline while controlling for baseline AMD status,24 and therefore encompassed progression from early to late AMD. Finally, some studies included additional covariates such as drusen size, smoking history, and BMI, which may alter the predictive power of genetic variants.
Similarly to what was seen in previous studies of both AMD disease risk and progression, we found a strong and significant effect of age2,24; older individuals progressed more rapidly to advanced AMD. However, because we did not know how long an eye had intermediate AMD before the first grade 3 exam, the effect of age encompasses an effect of disease duration. Older individuals may be more likely to have intermediate AMD for longer prior to their first grade 3 exam than younger individuals. Monitoring an individual's progression from unaffected control or grade 2 to an advanced case is necessary to fully determine the effect of age on AMD progression rate. Unlike risk of advanced AMD, which tends to be higher for females, AMD progression rate did not vary with sex as in several other studies.22,24,26
Considering progression to advanced AMD subtypes GA and CNV separately, the effect of CFH:rs10737680 was stronger and statistically significant only for predicting progression rate to CNV (HR = 1.83, P = 0.02 before Bonferroni correction). However, HR suggested that the effect was in a similar direction for progression to GA (HR = 1.56, P = 0.15; Table 3), and sample size of individuals who progressed to GA was slightly smaller (Supplementary Table S1). Similarly, when we considered the closely associated CFHR1-3 deletion-tagging SNP CFH:rs6677604, we found a similar effect to rs10737680, with a stronger association with progression to CNV compared to GA (see Results).
We found no difference in the effect of the other six variants on progression to either GA or CNV. A stronger effect of the rs10737680 proxy, rs1410996, on progression rate to CNV compared to GA was also found in the AREDS dataset, whereas two other CFH variants (Y402H and rs121913059) showed more similar effects on progression to either AMD form.26,33 However, in a separate study using the AREDS dataset, the two CFH variants (Y402H and rs2274700, a 100% proxy for rs10737680) showed similar effects on progression to GA and CNV in univariate analyses.28 Differences in genetic association between advanced AMD subtypes, including the variants tested here, have been reported for disease risk in case–control datasets, although overall there is high genetic correlation between the two subtypes.18 However, CFH variants are usually more strongly associated with increased risk of GA, and ARMS2 variants with CNV,17,18,53,54 whereas we found a stronger effect of CFH on progression to CNV. Nevertheless, growth rate of GA was correlated with genotype at the ARMS2 SNP in two studies,55,56 but not at the CFH (Y402H) SNP,55–57 although Caire et al.58 did not find evidence that GA growth rate was associated with variants in either locus. Overall, our results and previous work suggest that the CFH gene may be responsible for a slightly different AMD disease process than ARMS2,2,18 and that different variants may have different effects on risk versus progression. Factors that predict the incidence of advanced AMD may overlap more closely with those that predict whether or not progression to advanced AMD occurs, rather than progression rate.
This study replicated previous findings that rate of progression to advanced AMD is correlated with genotype at a common CFH variant, and that this association tended to be stronger for progression to CNV than to GA. This information may be used to identify individuals who are at increased risk of progressing quickly, thereby potentially improving monitoring and treatment. However, the effect of CFH on progression rate did not remain significant after correction for multiple tests, and its predictive power was not strong. Testing of association with other variants (both known and novel) and environmental factors such as smoking history and diet is important to accurately predict progression risk and to identify novel targets for drug development.
Supplementary Material
Acknowledgments
The authors thank all participants of this study.
Supported by National Eye Institute (NEI) Grant EY012118 (MAP-V, WKS, JLH, and AA), National Institutes of Health (NIH) Center Core Grant P30EY014801 (University of Miami), and an unrestricted grant from Research to Prevent Blindness, New York (University of Miami). Genotyping conducted as part of the International AMD Gene Consortium exome-chip project was supported by CIDR contract number HHSN268201200008I and funded by EY022310 (JLH) and 1X01HG006934-01 (to Gonçalo R. Abecasis). RJS and PJP were supported by a NEI T32 training grant (5T32EY023194-02).
Disclosure: R.J. Sardell, None; P.J. Persad, None; S.S. Pan, None; P. Whitehead, None; L.D. Adams, None; R.A. Laux, None; J.A. Fortun, Alcon (F), Allergan (C), DOR (C), Genentech (F); M.A. Brantley Jr, None; J.L. Kovach, None; S.G. Schwartz, None; A. Agarwal, None; J.L. Haines, None; W.K. Scott, None; M.A. Pericak-Vance, None
References
- 1. Klein R,, Chou C,, Klein BE,, Zhang X,, Meuer SM,, Saaddine JB. Prevalence of age-related macular degeneration in the US population. Arch Ophthalmol. 2011; 129: 75–80. [DOI] [PubMed] [Google Scholar]
- 2. Fritsche LG,, Fariss RN,, Stambolian D,, Abecasis GR,, Curcio CA,, Swaroop A. Age-related macular degeneration: genetics and biology coming together. Annu Rev Genomics Hum Genet. 2014; 15: 151–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Seddon JM,, Sharma S,, Adelman RA. Evaluation of the clinical age-related maculopathy staging system. Ophthalmology. 2006; 113: 260–266. [DOI] [PubMed] [Google Scholar]
- 4. Seddon JM,, Cote J,, Page WF,, Aggen SH,, Neale MC. The US twin study of age-related macular degeneration: relative roles of genetic and environmental influences. Arch Ophthalmol. 2005; 123: 321–327. [DOI] [PubMed] [Google Scholar]
- 5. Haines JL,, Hauser MA,, Schmidt S,, et al. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005; 308: 419–421. [DOI] [PubMed] [Google Scholar]
- 6. Hageman GS,, Anderson DH,, Johnson LV,, et al. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci U S A. 2005; 102: 7227–7232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Edwards AO,, Ritter R, III,, Abel KJ,, Manning A,, Panhuysen C,, Farrer LA. Complement factor H polymorphism and age-related macular degeneration. Science. 2005; 308: 421–424. [DOI] [PubMed] [Google Scholar]
- 8. Klein RJ,, Zeiss C,, Chew EY,, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005; 308: 385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Raychaudhuri S,, Iartchouk O,, Chin K,, et al. A rare penetrant mutation in CFH confers high risk of age-related macular degeneration. Nat Genet. 2011; 43: 1232–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gold B,, Merriam JE,, Zernant J,, et al. Variation in factor B (BF) and complement component 2 (C2) genes is associated with age-related macular degeneration. Nat Genet. 2006; 38: 458–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Maller J,, George S,, Purcell S,, et al. Common variation in three genes, including a noncoding variant in CFH strongly influences risk of age-related macular degeneration. Nat Genet. 2006; 38: 1055–1059. [DOI] [PubMed] [Google Scholar]
- 12. Maller JB,, Fagerness JA,, Reynolds RC,, Neale BM,, Daly MJ,, Seddon JM. Variation in complement factor 3 is associated with risk of age-related macular degeneration. Nat Genet. 2007; 39: 1200–1201. [DOI] [PubMed] [Google Scholar]
- 13. Yates JR,, Sepp T,, Matharu BK,, et al. Complement C3 variant and the risk of age-related macular degeneration. N Engl J Med. 2007; 357: 553–561. [DOI] [PubMed] [Google Scholar]
- 14. Rivera A,, Fisher SA,, Fritsche LG,, et al. Hypothetical LOC387715 is a second major susceptibility gene for age-related macular degeneration, contributing independently of complement factor H to disease risk. Hum Mol Genet. 2005; 14: 3227–3236. [DOI] [PubMed] [Google Scholar]
- 15. Yang Z,, Camp NJ,, Sun H,, et al. A variant of the HTRA1 gene increases susceptibility to age-related macular degeneration. Science. 2006; 314: 992–993. [DOI] [PubMed] [Google Scholar]
- 16. Jakobsdottir J,, Conley YP,, Weeks DE,, Mah TS,, Ferrell RE,, Gorin MB. Susceptibility genes for age-related maculopathy on chromosome 10q26. Am J Hum Genet. 2005; 77: 389–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fritsche LG,, Chen W,, Schu M,, et al. Seven new loci associated with age-related macular degeneration. Nat Genet. 2013; 45: 433–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fritsche LG,, Igl W,, Bailey JNC,, et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet. 2016; 48: 134–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Klein BE,, Klein R. Lifestyle exposures and eye diseases in adults. Am J Ophthalmol. 2007; 144: 961–969, e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Age-Related Eye Disease Study Research Group. Risk factors associated with age-related macular degeneration. A case-control study in the age-related eye disease study: Age-Related Eye Disease Study Report Number 3. Ophthalmology. 2000; 107: 2224–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tikellis G,, Robman L,, Dimitrov P,, Nicolas C,, McCarty C,, Guymer R. Characteristics of progression of early age-related macular degeneration: the Cardiovascular Health and Age-Related Maculopathy Study. Eye. 2006; 21: 169–176. [DOI] [PubMed] [Google Scholar]
- 22. Seddon JM,, Francis PJ,, George S,, Schultz DW,, Rosner B,, Klein ML. Association of CFH Y402H and LOC387715 A69S with progression of age-related macular degeneration. JAMA. 2007; 297: 1793–1800. [DOI] [PubMed] [Google Scholar]
- 23. Seddon JM,, Reynolds R,, Maller J,, Fagerness JA,, Daly MJ,, Rosner B. Prediction model for prevalence and incidence of advanced age-related macular degeneration based on genetic, demographic, and environmental variables. Invest Ophthalmol Vis Sci. 2009; 50: 2044–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Seddon JM,, Reynolds R,, Yu Y,, Daly MJ,, Rosner B. Risk models for progression to advanced age-related macular degeneration using demographic, environmental, genetic and ocular factors. Ophthalmology. 2011; 118: 2203–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gangnon RE,, Lee KE,, Klein BE,, Iyengar SK,, Sivakumaran TA,, Klein R. Effect of the Y402H variant in the complement factor H gene on the incidence and progression of age-related macular degeneration: results from multistate models applied to the Beaver Dam Eye Study. Arch Ophthalmol. 2012; 130: 1169–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yu Y,, Reynolds R,, Rosner B,, Daly MJ,, Seddon JM. Prospective assessment of genetic effects on progression to different stages of age-related macular degeneration using multistate Markov models. Invest Ophthalmol Vis Sci. 2012; 53: 1548–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gangnon RE,, Lee KE,, Klein BE,, Iyengar SK,, Sivakumaran TA,, Klein R. Severity of age-related macular degeneration in 1 eye and the incidence and progression of age-related macular degeneration in the fellow eye: the Beaver Dam Eye Study. JAMA Ophthalmol. 2015; 133: 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Perlee LT,, Bansal AT,, Gehrs K,, et al. Inclusion of genotype with fundus phenotype improves accuracy of predicting choroidal neovascularization and geographic atrophy. Ophthalmology. 2013; 120: 1880–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Baird PN,, Robman LD,, Richardson AJ,, et al. Gene-environment interaction in progression of AMD: the CFH gene smoking and exposure to chronic infection. Hum Mol Genet. 2008; 17: 1299–1305. [DOI] [PubMed] [Google Scholar]
- 30. Farwick A,, Wellmann J,, Stoll M,, Pauleikhoff D,, Hense HW. Susceptibility genes and progression in age-related maculopathy: a study of single eyes. Invest Ophthalmol Vis Sci. 2010; 51: 731–736. [DOI] [PubMed] [Google Scholar]
- 31. Francis PJ,, Hamon SC,, Ott J,, Weleber RG,, Klein ML. Polymorphisms in C2, CFB and C3 are associated with progression to advanced age related macular degeneration associated with visual loss. J Med Genet. 2009; 46: 300–307. [DOI] [PubMed] [Google Scholar]
- 32. Baird PN,, Richardson AJ,, Robman LD,, et al. Apolipoprotein (APOE) gene is associated with progression of age-related macular degeneration (AMD). Hum Mutat. 2006; 27: 337–342. [DOI] [PubMed] [Google Scholar]
- 33. Seddon JM,, Reynolds R,, Yu Y,, Rosner B. Three new genetic loci (R1210C in CFH variants in COL8A1 and RAD51B) are independently related to progression to advanced macular degeneration. PLoS One. 2014; 9: e87047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Klein R,, Knudtson MD,, Cruickshanks KJ,, Klein BE. Further observations on the association between smoking and the long-term incidence and progression of age-related macular degeneration: the Beaver Dam Eye Study. Arch Ophthalmol. 2008; 126: 115–121. [DOI] [PubMed] [Google Scholar]
- 35. Chew EY. Nutrition effects on ocular diseases in the aging eye. Invest Ophthalmol Vis Sci. 2013; 54: ORSF42–ORSF47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schmidt S,, Saunders AM,, De La Paz M,, et al. Association of the apolipoprotein E gene with age-related macular degeneration: possible effect modification by family history, age, and gender. Mol Vis. 2000; 6: 93. [PubMed] [Google Scholar]
- 37. 1000 Genomes Project Consortium. An integrated map of genetic variation from 1092 human genomes. Nature. 2012; 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. O'Connell J,, Gurdasani D,, Delaneau O,, et al. A general approach for haplotype phasing across the full spectrum of relatedness. PLoS Genet. 2014; 10: e1004234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Howie BN,, Donnelly P,, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009; 5: e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Price AL,, Patterson NJ,, Plenge RM,, Weinblatt ME,, Shadick NA,, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006; 38: 904–909. [DOI] [PubMed] [Google Scholar]
- 41. Purcell S,, Neale B,, Todd-Brown K,, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007; 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Therneau TM. A Package for Survival Analysis in S (2015). Version 2.38. Available at: https://CRAN.R-project.org/package=survival.
- 43. Pauleikhoff D,, Radermacher M,, Spital G,, et al. Visual prognosis of second eyes in patients with unilateral late exudative age-related macular degeneration. Graefes Arch Clin Exp Ophthalmol. 2002; 240: 539–542. [DOI] [PubMed] [Google Scholar]
- 44. Glynn RJ,, Rosner B. Methods to quantify the relation between disease progression in paired eyes. Am J Epidemiol. 2000; 151: 965–974. [DOI] [PubMed] [Google Scholar]
- 45. Heagerty PJ,, Lumley T,, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000; 56: 337–344. [DOI] [PubMed] [Google Scholar]
- 46. Owzar K,, Li Z,, Cox N,, Jung S. Power and sample size calculations for SNP association studies with censored time-to-event outcomes. Genet Epidemiol. 2012; 36: 538–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2015. Available at: http://www.R-project.org/. [Google Scholar]
- 48. Raychaudhuri S,, Ripke S,, Li M,, et al. Associations of CFHR1-CFHR3 deletion and a CFH SNP to age-related macular degeneration are not independent. Nat Genet. 2010; 42: 553–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fritsche LG,, Lauer N,, Hartmann A,, et al. An imbalance of human complement regulatory proteins CFHR1, CFHR3 and factor H influences risk for age-related macular degeneration (AMD). Hum Mol Genet. 2010; 19: 4694–4704. [DOI] [PubMed] [Google Scholar]
- 50. Ansari M,, McKeigue PM,, Skerka C,, et al. Genetic influences on plasma CFH and CFHR1 concentrations and their role in susceptibility to age-related macular degeneration. Hum Mol Genet. 2013; 22: 4857–4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hughes AE,, Orr N,, Esfandiary H,, Diaz-Torres M,, Goodship T,, Chakravarthy U. A common CFH haplotype, with deletion of CFHR1 and CFHR3 is associated with lower risk of age-related macular degeneration. Nat Genet. 2006; 38: 1173–1177. [DOI] [PubMed] [Google Scholar]
- 52. Seddon JM,, Reynolds R,, Yu Y,, Rosner B. Validation of a prediction algorithm for progression to advanced macular degeneration subtypes. JAMA Ophthalmol. 2013; 131: 448–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chen W,, Stambolian D,, Edwards AO,, et al. Genetic variants near TIMP3 and high-density lipoprotein-associated loci influence susceptibility to age-related macular degeneration. Proc Natl Acad Sci U S A. 2010; 107: 7401–7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sobrin L,, Reynolds R,, Yu Y,, et al. ARMS2/HTRA1 locus can confer differential susceptibility to the advanced subtypes of age-related macular degeneration. Am J Ophthalmol. 2011; 151: 345–352, e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Grassmann F,, Fleckenstein M,, Chew EY,, et al. Clinical and genetic factors associated with progression of geographic atrophy lesions in age-related macular degeneration. PLoS One. 2015; 10: e012663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Klein ML,, Ferris FL, III,, Francis PJ,, et al. Progression of geographic atrophy and genotype in age-related macular degeneration. Ophthalmology. 2010; 117: 1554–1559, e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Scholl HP,, Fleckenstein M,, Fritsche LG,, et al. CFH C3 and ARMS2 are significant risk loci for susceptibility but not for disease progression of geographic atrophy due to AMD. PLoS One. 2009; 4: e7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Caire J,, Recalde S,, Velazquez-Villoria A,, et al. Growth of geographic atrophy on fundus autofluorescence and polymorphisms of CFH, CFB, C3, FHR1-3, and ARMS2 in age-related macular degeneration. JAMA Ophthalmol. 2014; 132: 528–534. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.