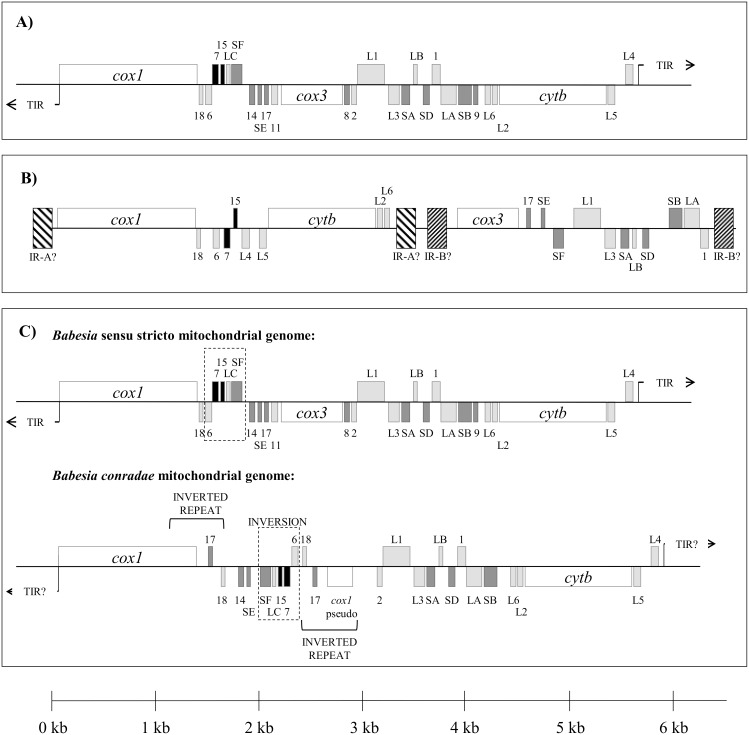
Fig 2. Mitochondrial genome structures of Piroplasmida species characterized in this study.
Genes shown above the central line are coded on the sense strand, while those below are on the antisense strand. Protein-coding genes (cox1, cox3, and cytb) are indicated in white. Large subunit rRNA fragments are in light gray, small subunit rRNA fragments are in dark gray, and miscellaneous conserved RNA fragments are in black. A) Mitochondrial genome sequences of C. felis, B. rossi, B. vogeli, B. canis, and Babesia sp. Coco maintained the mitochondrial genome structure that is characteristic of traditional Babesia sensu stricto and Theileria species, while B) the inferred Babesia microti-like sp. mitochondrial genome structure appears to be similar to that of B. microti and B. rodhaini, suggesting it has a “flip-flop” mitochondrial genome structure. Assumed inverted repeats A and B (indicated as IR-A and IR-B) were not confirmed due to lack of relevant sequence for phylogenetic analysis. C) Babesia conradae had a unique mitochondrial genome, which lacked cox3 and had a duplicated inversion that included the 3’ end of cox1 and RNA17 and RNA18. Additionally, a collection of rRNA fragments (RNA6, RNA7, RNA15, LSUC, and SSUF) found in Babesia sensu stricto, C. felis, and Theileria mitochondrial genomes was conserved but inverted as a unit adjacent to the duplicated inversion.