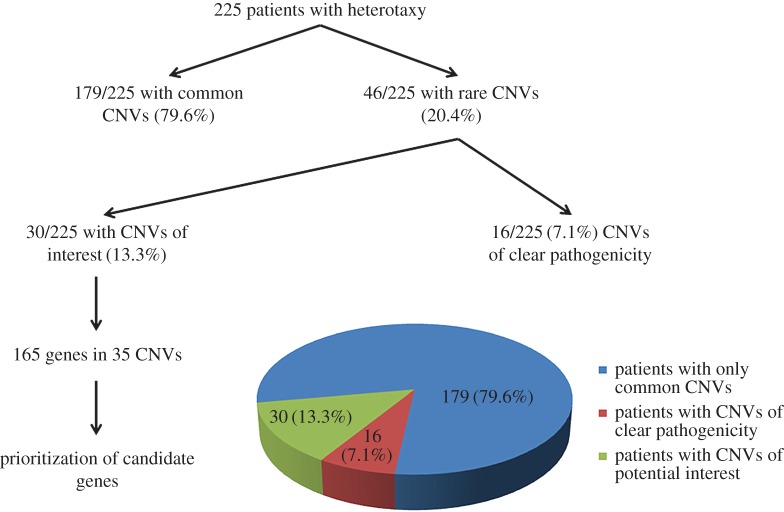
Figure 1.
Overview of CNV findings. CNVs were designated as ‘common’ or ‘rare’ based on their presence in clinical and online databases of genomic variation (see §2). CNVs denoted as having ‘clear pathogenicity’ included a diverse array of large/complex chromosomal abnormalities (table 3), as well as CNVs containing genes previously associated with heterotaxy (table 4). CNVs ‘of interest’ represent CNVs containing potential candidate genes, including some identified at loci previously associated with CHD (electronic supplementary material, table S3) or with microduplication/deletion syndromes (electronic supplementary material, table S4).