Figure 5.
Mitochondrial Dynamics Regulate Cell Size Scaling of Mitochondrial Functions
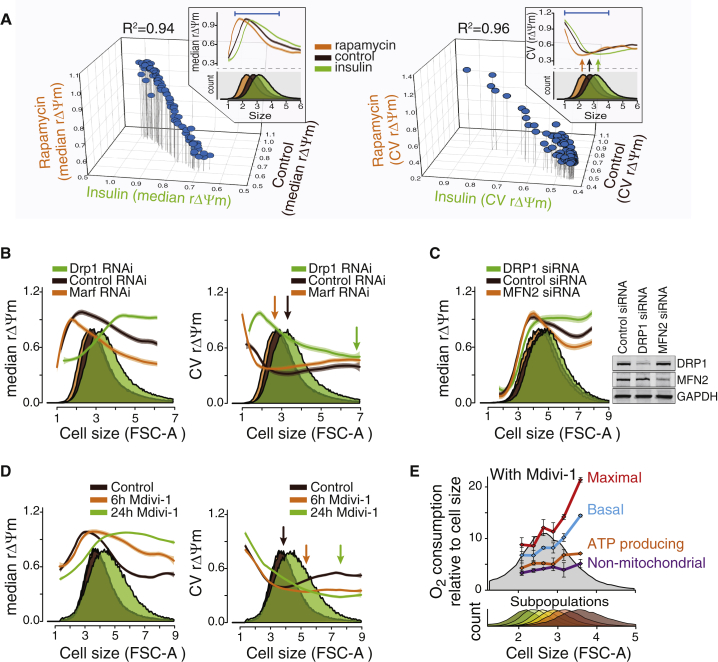
(A) Insulin signaling controls optimal cell size without changing the scaling of rΔΨm. Kc167 cells were treated with insulin and rapamycin for 14 hr and rΔΨm was analyzed with JC-1 staining. The 3D plots display median (left) and variability (right) of rΔΨm for each cell size bin after normalizing for the overall cell size change. Insets display size distribution and scaling of rΔΨm and its variability. n > 2 × 105 cells.
(B) Cell size scaling of rΔΨm (left) and its cell-to-cell variability (right) in Kc167 cells after 5 days of double-stranded RNA-mediated knockdown of Drp1 and Marf (mitofusin). Arrows indicate the cell size with the minimum variability. n > 1.7 × 105 cells.
(C) Cell size scaling of rΔΨm in Jurkat cells after 65 hr of small interfering RNA-mediated knockdown of DRP1 and MNF2 (mitofusin 2). Inset displays western blot of the knockdown efficiency. n > 1.2 × 105 cells.
(D) Kc167 cell rΔΨm (left) and variability (right) changes after 6 and 24 hr of treatment with the Drp1 inhibitor Mdivi-1. Arrows indicate the cell size with the minimum variability. n > 2.5 × 105 cells.
(E) The changes in the rΔΨm caused by Mdivi-1 translate into increased oxygen consumption in the largest cells. Kc167 cells treated with Mdivi-1 were separated by centrifugal elutriation to size-based subpopulations (bottom panel) from which oxygen consumption parameters were analyzed. Oxygen consumption data represent mean ± SD (n = 3–4).
See also Figure S5.