Abstract
Hemolysis is a complication in septic infections with Staphylococcus aureus, which utilizes the released Hb as an iron source. S. aureus can acquire heme in vitro from hemoglobin (Hb) by a heme-sequestering mechanism that involves proteins from the S. aureus iron-regulated surface determinant (Isd) system. However, the host has its own mechanism to recapture the free Hb via haptoglobin (Hp) binding and uptake of Hb-Hp by the CD163 receptor in macrophages. It has so far remained unclear how the Isd system competes with this host iron recycling system in situ to obtain the important nutrient. By binding and uptake studies, we now show that the IsdH protein, which serves as an Hb receptor in the Isd system, directly interferes with the CD163-mediated clearance by binding the Hb-Hp complex and inhibiting CD163 recognition. Analysis of truncated IsdH variants including one or more of three near iron transporter domains, IsdHN1, IsdHN2, and IsdHN3, revealed that Hb binding of IsdHN1 and IsdHN2 accounted for the high affinity for Hb-Hp complexes. The third near iron transporter domain, IsdHN3, exhibited redox-dependent heme extraction, when Hb in the Hb-Hp complex was in the oxidized met form but not in the reduced oxy form. IsdB, the other S. aureus Hb receptor, failed to extract heme from Hb-Hp, and it was a poor competitor for Hb-Hp binding to CD163. This indicates that Hb recognition by IsdH, but not by IsdB, sterically inhibits the receptor recognition of Hb-Hp. This function of IsdH may have an overall stimulatory effect on S. aureus heme acquisition and growth.
Keywords: cell surface receptor, hemoglobin, iron, macrophage, Staphylococcus aureus (S. aureus), CD163, Iron-regulated Surface Determinant, IsdH, haptoglobin, heme acquisition
Introduction
Staphylococcus aureus is a Gram-positive bacterium that colonizes approximately one-third of the human population (1). It can be invasive and cause an array of diseases including hemolysis and septic shock. Successful host invasion involves compromising the efficacy of the immune system and efficient acquisition of essential nutrients including iron. Like a number of other pathogenic bacteria (e.g. strains of Escherichia coli, Pseudomonas, and Streptococci) (2–4), S. aureus secretes an α-hemolysin that integrates in red blood cell membranes and induces osmotic hemolysis. Liberation of Hb into plasma facilitates S. aureus acquisition of iron by means of an iron-sequestering pathway designated the iron-regulated surface determinant (Isd)3 system (5, 6). S. aureus expresses several different Isd proteins (IsdA, IsdB, IsdC, IsdE, IsdF, IsdG, and IsdH) that orchestrate the acquisition of host Hb heme iron. The functions of most of these proteins have been elucidated: extraction of heme is achieved by the two bacterial surface-exposed Hb-receptors, IsdB and IsdH; transport of heme across the bacterial cell wall and plasma membrane is performed by IsdA and IsdC together with the membrane protein IsdEF complex; and the heme oxygenase enzymes IsdG and IsdH, located in the cytoplasm, finally cleave the porphyrin ring (reviewed in Ref. 7). Although the role of Isd proteins in the sequestering of iron from free Hb is well understood, this may not apply to the situation in the blood where extracellular Hb is found in complex with Hp.
The heme-binding function of S. aureus Isd proteins—IsdA, IsdB, IsdC, and IsdH—is conferred by the presence of a near iron transporter (NEAT) domain with a conserved heme-binding pocket (8). Importantly, the heme-binding domain alone is unable to extract heme from Hb, and IsdB and IsdH contain additional NEAT domains to achieve this function (9). Thus, IsdH contains three NEAT domains of which the first and second NEAT domain (IsdHN1 and IsdHN2) bind to Hb but lack heme binding activity, whereas the third, C-terminal, NEAT domain (IsdHN3) carries the single heme-binding site of IsdH. IsdHN2 and IsdHN3 are connected by an α-helical linker domain and the IsdHN2-linker-IsdHN3 region is the minimal fragment of the IsdH receptor that retains native ability to capture heme from Hb (9–11). IsdB has a two-NEAT domain (IsdBN1 and IsdBN2) structure connected with an α-helical linker domain, similar to the minimal functional fragment of IsdH (8). In addition to Hb binding, the IsdHN1 domain is also reported to bind other ligands including Hp (12, 13). Independent of heme extraction, IsdH also plays a role in S. aureus immune evasion by promoting degradation of bound complement C3, thereby avoiding opsonophagocytosis (14).
Hb released into human plasma during hemolysis binds rapidly to plasma Hp, which protects against the highly oxidative and toxic properties of Hb by direct shielding of oxidative spots (15, 16) and by the promotion of Hb-uptake via the macrophage-specific endocytic receptor CD163 (17–21). Hp exists in three main variants designated Hp1-1, Hp2–1, and Hp2-2, where Hp1-1 is a Hp dimer, whereas the two other variants are found as different multimeric forms. All forms bind αβ-Hb dimers in the Hp region distal to the center of the Hp protein. Structural data have shown that IsdH binds to Hb in Hb-Hp complexes close to the site for interaction of Hp and CD163 (22).
In the present study, we show that IsdH only binds Hb-Hp via a direct Hb interaction without direct contact to the Hp subunit, in contrast to previous reporting (12, 13, 23) of a direct low affinity interaction between IsdH and Hp. Furthermore, this study describes how the interaction with Hb-Hp leads to heme extraction and obstruction of CD163-mediated clearance of Hb-Hp complexes in macrophages.
Results
Binding of IsdH to Hb-Hp Complexes
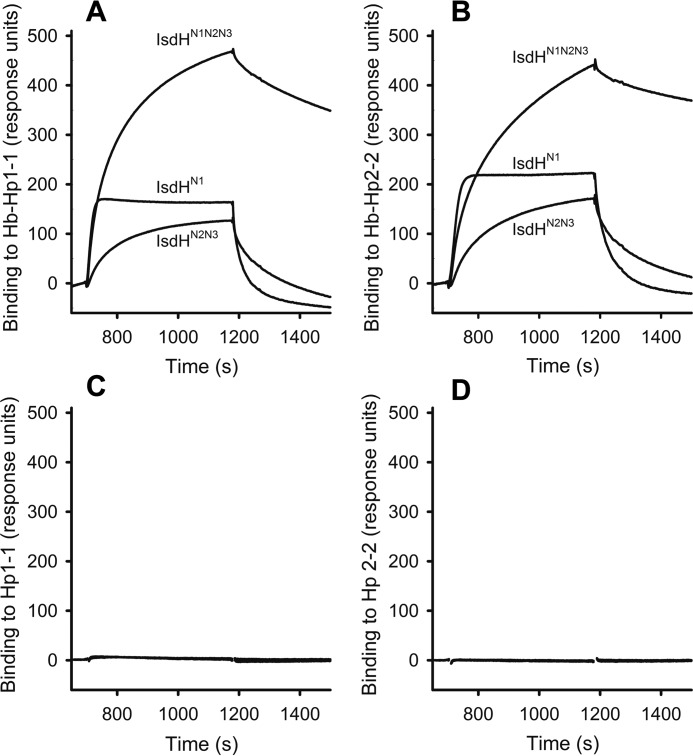
IsdH truncation variants IsdHN1, IsdHN2N3, IsdHN1N2N3, and IsdBN1N2 with a His6 tag were expressed recombinantly and analyzed by SDS-PAGE (Fig. 1). Binding of the IsdH variants to Hb-Hp complexes was evaluated by SPR analysis (Fig. 2, A and B). The apparent equilibrium dissociation constants for binding of IsdHN1 (one Hb binding site), IsdHN2N3 (one Hb binding site), or IsdHN1N2N3 (two Hb binding sites) to Hb-Hp1-1 or Hp2-2 were ∼70,∼120, and ∼30 nm, respectively (Fig. 2). The binding curves of IsdHN1 and IsdHN1N2N3 were different in the sense that IsdHN1 and IsdHN2N3 displayed faster off rates, compared with IsdHN1N2N3. These results suggest that the two Hb-binding domains in IsdHN1 and IsdHN2, together increase the overall functional affinity of IsdHn1N2N3 for Hb-Hp. No significant binding of any of the IsdH constructs was seen to Hp1-1 and Hp2-2 (Fig. 2, C and D).
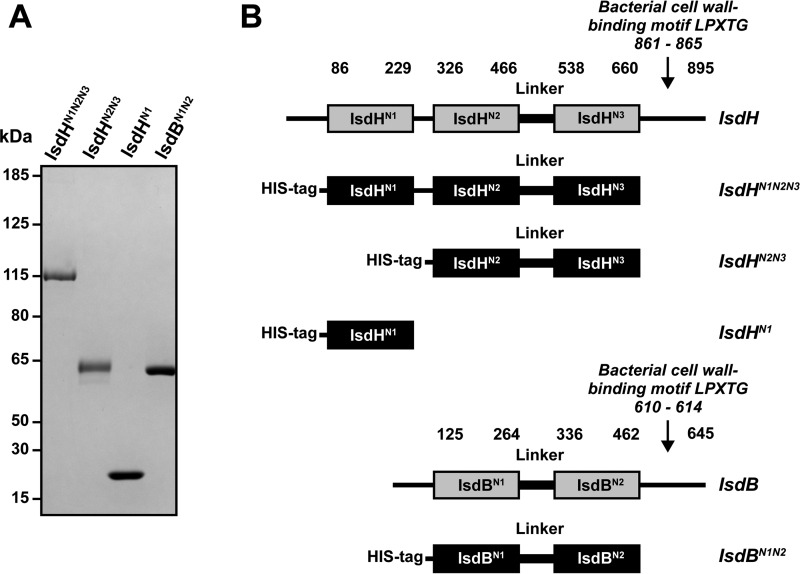
FIGURE 1.
SDS-PAGE of IsdH constructs. A, IsdHN1, IsdHN2N3, IsdHN1N2N3, and IsdBN1N2 were expressed recombinantly in E. coli with N-terminal His6 tags, purified, and analyzed by SDS-PAGE. B, a schematic overview of full-length IsdH, IsdB, and the derived constructs used in this study. Full-length IsdH consists of three NEAT domains and a linker connecting the second and third NEAT domain. Full-length IsdB consists of two NEAT domains connected with a linker. An LPXTG sequence in C-terminal part links IsdH and IsdB to the bacterial cell wall (40). The truncated variants IsdHN1N2N3 (residues 82–655), IsdHN2N3 (residues 321–655), IsdHN1 (residues 86–229), and IsdBN1N2 (residues 120–459) were used in this study.
FIGURE 2.
SPR analysis of IsdH binding to Hb-Hp. A and B, binding of 100 nm IsdHN1, IsdHN2N3, and IsdHN1N2N3 to immobilized Hp1-1 saturated with Hb (A) or immobilized Hp2-2 saturated with Hb (B). On basis of these data, the apparent KD values were estimated by fitting the plateau binding response: IsdHN1 binding to Hb-Hp1-1 KD = 74 ± 18 nm and binding to Hb-Hp2-2 KD = 71 ± 19 nm; IsdHN2N3 binding to Hb-Hp1-1 KD = 117 ± 17 nm and binding to Hb-Hp2-2 KD = 127 ± 18 nm; IsdHN1N2N3 binding to Hb-Hp1-1 KD = 28 ± 10 nm and to Hb-Hp2-2 KD = 33 ± 9 nm. The values are represented as means ± S.D., and r2 of the fits were all above 0.98. C and D, binding of 100 nm IsdHN1, IsdHN2N3, and IsdHN1N2N3 to immobilized Hp1-1 (C) or immobilized Hp2-2 (D). The binding was investigated on two independently produced flow cells for both Hp1-1 and Hp2-2, respectively. All experiments were at least repeated in triplicate to each flow cell. The apparent binding constants are determined based on a triplicate binding experiment to a single flow cell with either Hp1-1 or Hp2-2.
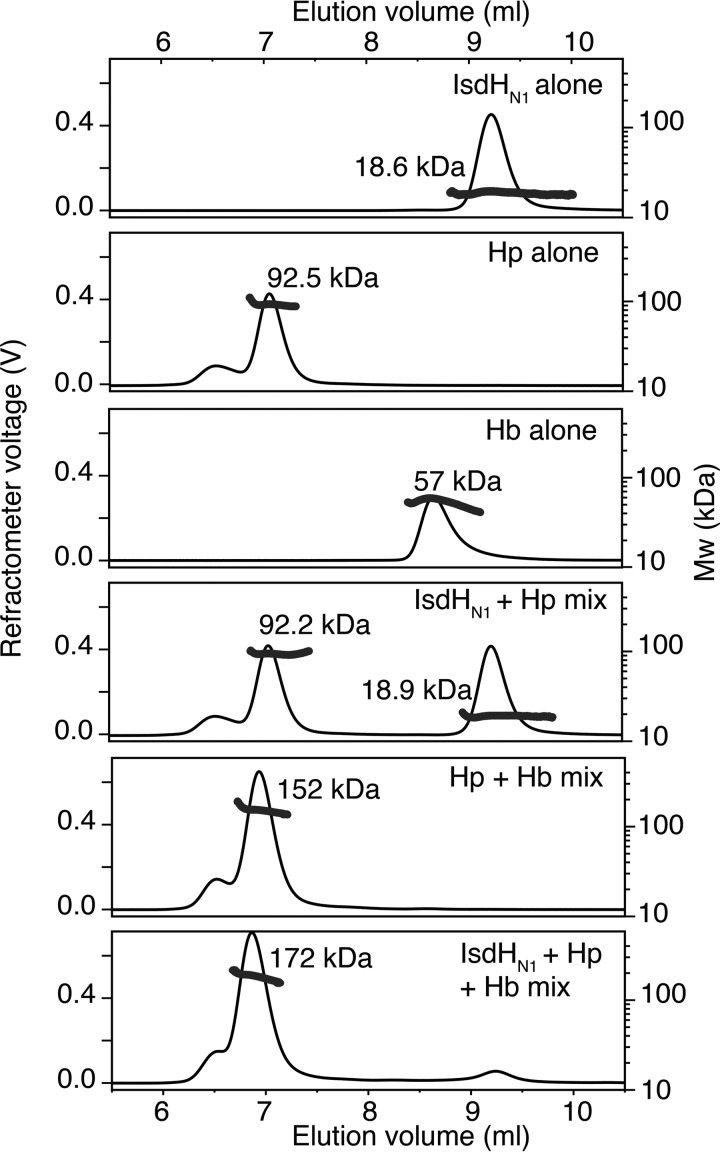
To further characterize the interaction between IsdHN1 and Hb or Hb-Hp complexes in solution, we employed size exclusion chromatography (SEC) with in-line right angle light scattering (RALS). The molecular mass of IsdHN1 determined by SEC-RALS in solution (18.6 kDa) was close to the monomer molecular mass based on sequence and determined by mass spectrometry (18,907 Da) (Fig. 3). The major component of the Hp1-1 preparation was a ∼93-kDa species, consistent with the presence of the Hp1-1 dimer, including ∼20% mass contribution because of glycosylation; a minor peak at the exclusion volume of the SEC column was assumed to be an aggregate. Hb eluted as a single asymmetric peak, indicative of the well characterized dimer-tetramer equilibrium (24); the measured molecular mass of 57 kDa suggested that the protein was predominantly in the tetrameric state under the experimental conditions. Analysis of a 1:1 (by mass) mixture of IsdHN1 with Hp showed no significant change in the peak elution times or molecular masses as determined by RALS (Fig. 3); thus no interaction between IsdHN1 and Hp could be detected. In contrast, Hp and Hb in an 1:1 (by mass) mixture formed a complex with a molecular mass of 152 kDa, consistent with a stoichiometry of one Hp1-1 dimer bound to two Hb αβ-dimers; the absence of any detectable free Hb is consistent with the known high affinity binding between Hp and Hb. The elution of this complex shifted to an earlier time point, and the molecular mass of the complex increased upon addition of IsdHN1 (Fig. 3), indicating formation of a ternary Hb-Hp-IsdHN1 complex. Taken together, the SPR and SEC-RALS data show that Hb is necessary for interaction between Hb-Hp and IsdH, most likely via a direct interaction between IsdH and Hb.
FIGURE 3.
SEC-LS analysis of IsdHN1 mixtures with Hp or Hb-Hp. Elution profiles are shown from 50-μl injections of IsdHN1, Hp1-1, or Hb at 1 mg/ml or mixtures of IsdHN1 + Hp1-1 (1 mg/ml each), Hp1-1 + Hb (1 mg/ml each), and IsdHN1 + Hp1-1 + Hb (0.4, 1, and 1 mg/ml respectively). Refractometer voltage (solid line) is proportional to mass concentration. Molecular mass was calculated from multiple refractive index and light scattering measurements across each peak (solid data points), and the molecular mass calculated across the whole peak is shown.
IsdH Acquisition of Heme from Hb-Hp
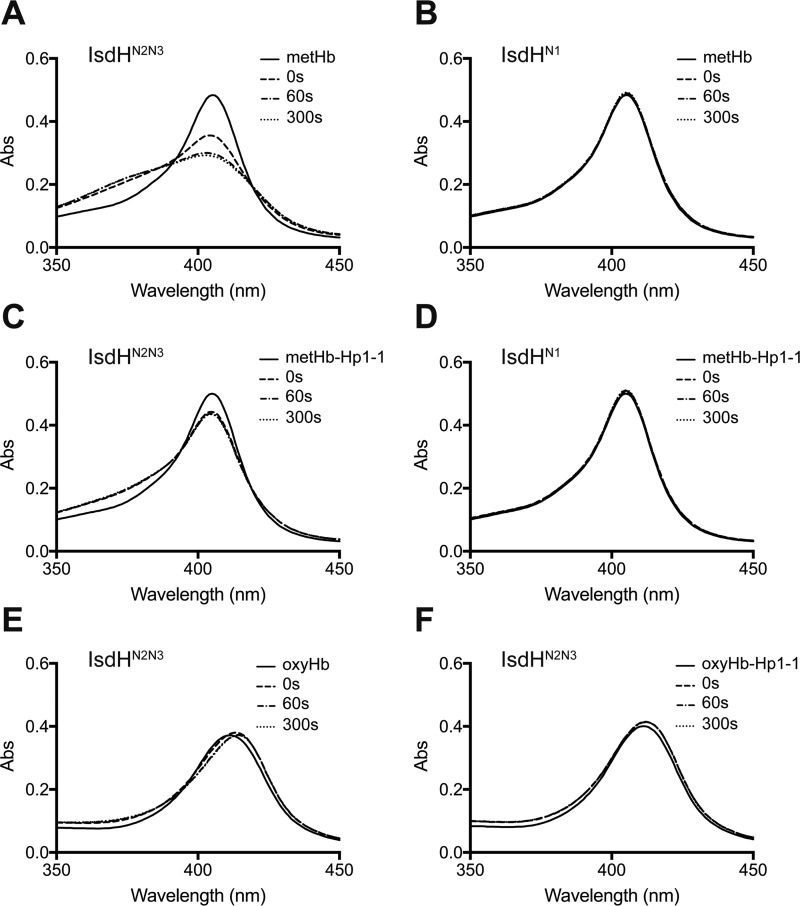
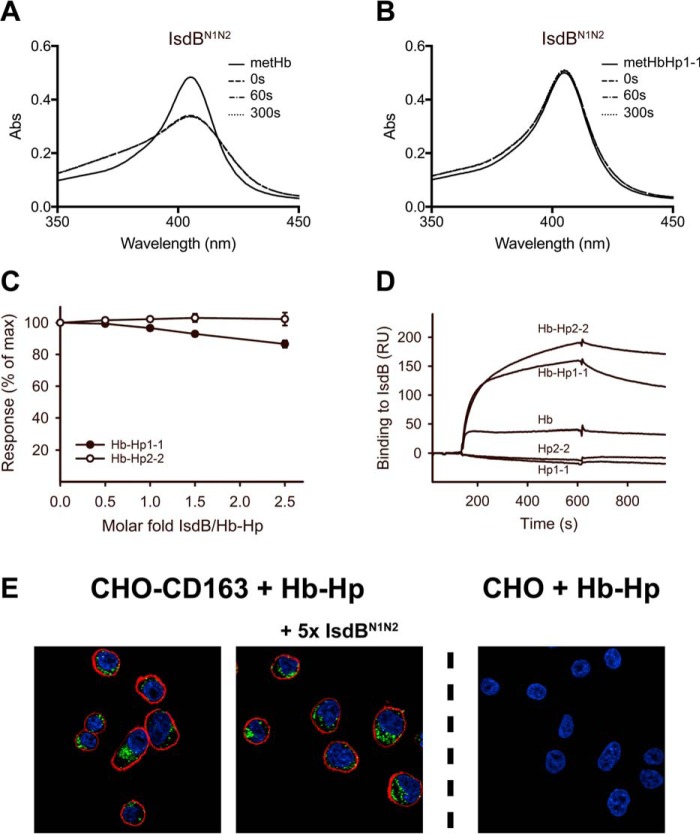
To determine whether IsdHN3 is capable of extracting heme from Hb-Hp, heme transfer from Hb-Hp to IsdHN2N3 was monitored using UV-visible spectroscopy, taking advantage of the different spectra generated when heme is bound to the globin or bound to the IsdH protein. Previous studies have confirmed rapid heme transfer to IsdHN2N3 from metHb (9, 13). Similar results were obtained when metHb was incubated in the presence of added IsdHN2N3 (Fig. 4A). Significant absorbance changes were also evident when metHb-Hp1-1 was incubated with IsdHN2N3 (Fig. 4C), indicating that IsdH is capable of extracting heme from Hb in complex with Hp. When IsdHN2N3 was mixed with ferrous oxyHb or oxyHb-Hp1-1, no spectral changes were detected (Fig. 4, E and F). Thus, ferrous oxyHb has to be oxidized to metHb for IsdH to be able to extract heme.
FIGURE 4.
IsdH-mediated heme transfer from Hb and Hb-Hp. A–F, spectral changes over time for 1.5 μm metHb (tetramer) mixed with 13 μm IsdHN2N3 (A) or 13 μm IsdHN1 (which cannot extract heme) (B), 1.5 μm metHb-Hp1-1 (Hp dimer; two Hb dimers) mixed with 13 μm IsdHN2N3 (C) or 13 μm IsdHN1 (D), 1.5 μm oxyHb (tetramer) mixed with 13 μm IsdHN2N3 (E), and 1.5 μm oxyHb-Hp1-1 (Hp dimer; two Hb dimers) mixed with 13 μm IsdHN2N3 (F). The time points 0, 60, and 300 s indicate time after the first recording. For comparison, 1.5 μm metHb or 1.5 μm metHb-Hp1-1 mixed 1:1 with PBS are included (solid line). By estimating from the absorbance changes only, 39.6 ± 0.2% and 12.8 ± 1.7% (n = 3) of heme was transferred to IsdHN2N3 in 300 s from metHb and metHb-Hp1-1, respectively. No measurable transfer to IsdHN1 was seen in this time span.
IsdH Interference with CD163 Binding of Hb-Hp
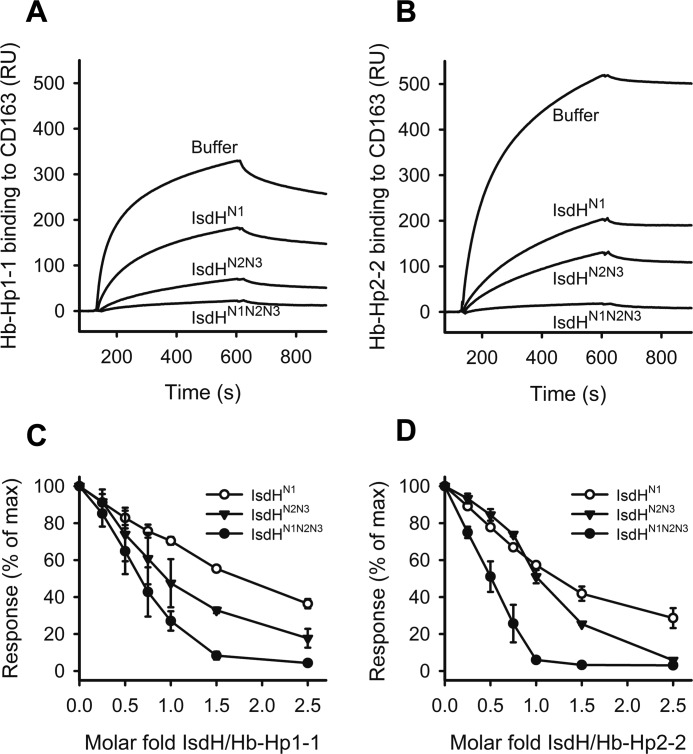
Surface plasmon resonance (SPR) analysis was used to determine whether the high affinity binding of IsdH to Hb-Hp interferes with the binding of the complex to purified CD163. A robust SPR response was obtained when Hb-Hp1-1 was injected over CD163 immobilized on the SPR chip surface (Fig. 5A, Buffer), indicating a complex forming between Hb-Hp1-1 and CD163. In contrast, when Hb-Hp1-1 was injected together with an equimolar concentration of IsdHN1, the SPR response was strongly attenuated (Fig. 5A), suggesting that IsdHN1 interferes with the interaction of Hb-Hp1-1 with CD163. Similarly, the IsdHN2N3 and IsdHN1N2N3 proteins also blocked the Hb-Hp1-1 interactions with CD163 (Fig. 5, A and C). Similar results were obtained for interactions of Hb-Hp2-2 with CD163 (Fig. 5, B and D). All IsdH constructs exhibited dose-dependent inhibition of Hb-Hp binding to CD163 with IsdHN1N2N3 showing the highest potency. The IsdH variants were slightly more potent in their inhibition of Hb-Hp2-2 (Fig. 5, B and D) binding compared with Hb-Hp1-1 (Fig. 5, A and C).
FIGURE 5.
SPR analysis of CD163-mediated binding of Hb-Hp in presence of IsdH. A and B, sensorgrams displaying binding of Hb-Hp1-1 (A) and Hb-Hp2-2 (B) to immobilized CD163 in presence of 1.5 molar equivalent of IsdHN1, IsdHN2N3, and IsdHN1N2N3 or running buffer as control. C and D, relative plateau response of immobilized CD163 binding of Hb-Hp1-1 (C) and Hb-Hp2-2 (D) are plotted against the indicated molar ratio of IsdHN1 (○), IsdHN2N3 (▾), or IsdHN1N2N3(●). Reproduction of the SPR experiments was ensured by preparing two individual chips with two individual flow cells with either Hp1-1 or Hp2-2. On both chips triplicate repeats of triplicate runs of each sample was performed. The shown data are representative sensorgrams (A and B) and representative results from triplicate experiments to a single flow cell (C and D).
To investigate how these interactions may impact on clearance of Hb-Hp complexes by cellular CD163, we analyzed the endocytosis of fluorophore-labeled Hb-Hp2-2 by transfected CD163-expressing CHO cells (CHO CD163) in the presence of IsdHN1N2N3, IsdHN2N3, or IsdHN1. As expected, confocal microscopy showed that fluorophore-labeled Hb-Hp (Fig. 6A, green) only was endocytosed when CHO cells were expressing CD163 (Fig. 6A, red). A substantial inhibition of CD163-mediated vesicular Hb-Hp uptake was observed in the presence of IsdHN1N2N3, IsdHN2N3, or IsdHN1 compared with the positive control (Fig. 6A). The inhibitory effect by IsdH was quantified by analyzing the cellular uptake of fluorescent Hb-Hp2-2 by flow cytometry (Fig. 6B). All IsdH constructs inhibited Hb-Hp uptake in a dose-dependent manner, with IsdHN1N2N3 being most efficient showing a 6-fold reduction in uptake using an equimolar concentration of the Hb-Hp units. Complete inhibition, corresponding to the background level in the mock transfected CHO cells, was seen at 5 molar excess of IsdHN1N2N3 and IsdHN1. The inhibitory effect by IsdH was also evident in flow cytometric analysis of monocytes/macrophages, the cell type responsible for the in vivo clearance of Hb-Hp (Fig. 7). The flow cytometric analysis shows that an equimolar concentration of IsdHN1N2N3 reduced the mean fluorescence intensity (MFI) signal of Hb-Hp ∼4-fold. Similar inhibition was a seen with IsdHN1 and IsdHN2N3, although 5-fold higher concentrations of these IsdH truncation mutants were needed to obtain the same level of inhibition.
FIGURE 6.
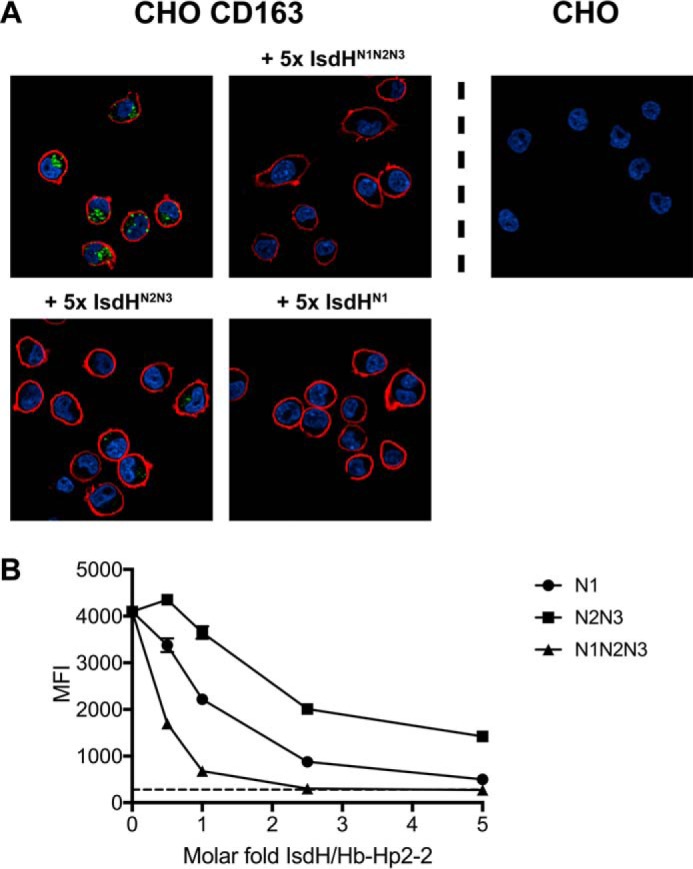
Analysis of cellular uptake of Hb-Hp2-2 by CD163-expressing cells. A, confocal microscopy images of endocytotic Hb-Hp uptake by CD163-expressing cells. CHO CD163 cells were incubated with 46 μg/ml Atto-488-labeled Hb-Hp2-2 and IsdHN1, IsdHN2N3, or IsdHN1N2N3 in 5-fold molar excess over Hb-Hp. Atto-488-labeled Hb-Hp2-2 is shown in green, DAPI staining of nuclei is shown in blue, and CD163 surface immunostaining (Alexa 647) is shown in red. Mock transfected CHO cells (right) were included as a negative control. B, dose-response inhibition of endocytotic uptake of Atto-488 labeled Hb-Hp2-2 by CHO CD163 cells in presences of increasing molar excess of IsdHN1 (○), IsdHN2N3 (■), or IsdHN1N2N3 (▴). Fluorescence signals were detected by flow cytometry and displayed as MFI. Background uptake in mock transfected CHO cells is indicated with a dashed line. The data are represented as means ± S.D. (n = 3) from one representative experiment of three.
FIGURE 7.
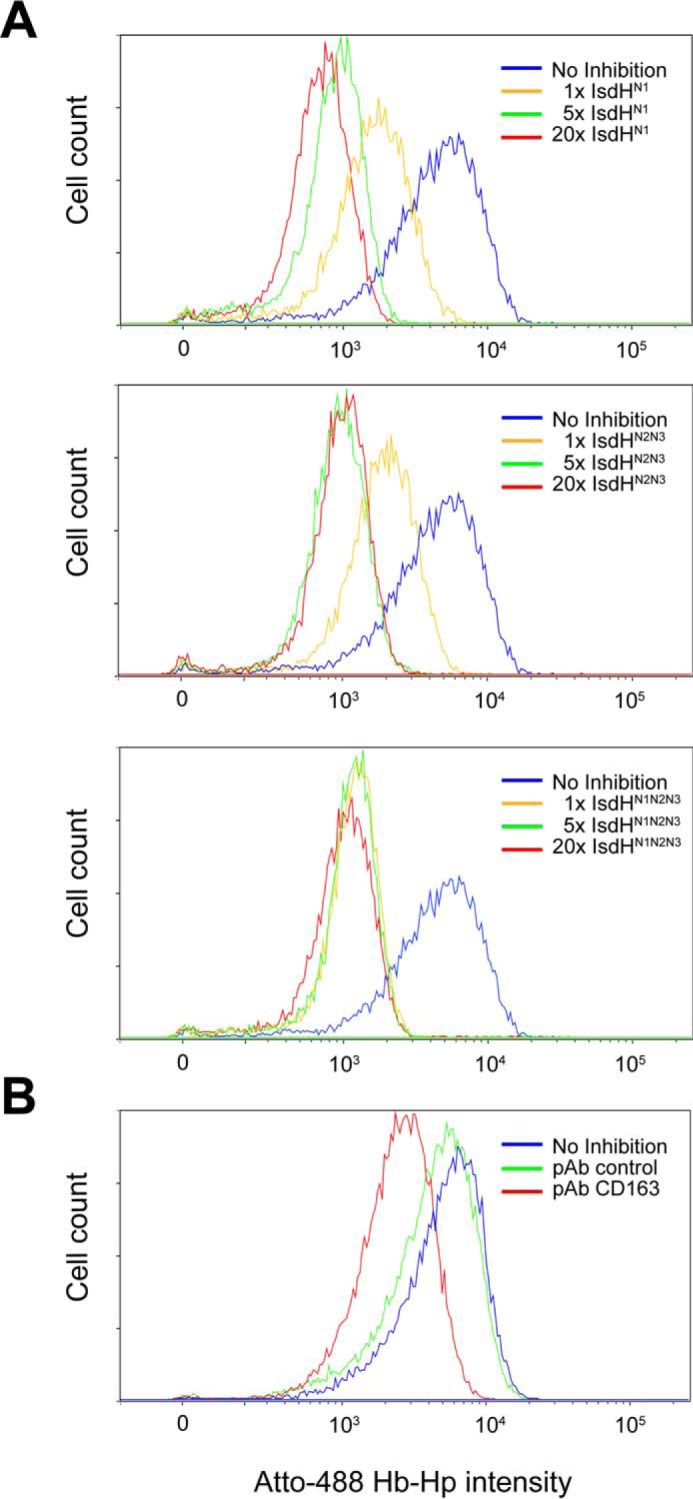
Flow cytometric analysis of uptake of Hb-Hp2-2 by dexamethasone-treated monocytes. A, adherent 3-day-old monocytes treated with 2.5 × 10−7 m dexamethasone were incubated with 23 μg/ml Atto-488-labeled Hb-Hp2-2 and IsdHN1, IsdHN2N3, or IsdHN1N2N3 in 1-, 5-, and 20-fold molar excess over Hb-Hp. B, a polyclonal rabbit anti-human CD163 IgG (50 μg/ml) and an irrelevant polyclonal rabbit anti-human antibody (50 μg/ml) were used as positive and negative inhibition controls respectively. The data from one representative experiment of three are shown.
Comparative Analyses of IsdB
Next, because IsdH and IsdB share a high degree of structural homology and both are reported to function as Hb receptors, interactions of IsdB with the Hb-Hp complex and its removal by CD163 were investigated. IsdBN1N2 was analyzed by UV-visible spectroscopy for its ability to transfer heme (Fig. 8, A and B). Although IsdBN1N2 rapidly transfers heme from metHb, we could not detect any heme transfer from metHb-Hp. In contrast to IsdH, SPR analysis showed no or very weak inhibition of Hb-Hp binding to CD163 by IsdBN1N2 (Fig. 8C), although IsdBN1N2 was able to bind to Hb-Hp (Fig. 8D). Finally, IsdBN1N2 did not inhibit uptake of fluorophore-labeled Hb-Hp2-2 in CHO CD163 (Fig. 8E).
FIGURE 8.
IsdB heme transfer kinetics, SPR analysis, and cellular uptake of Hb-Hp2-2. A and B, spectral changes over time after mixing of 13 μm sIsdBN1N2 with 1.5 μm metHb (tetramer) (A) or 1.5 μm metHb-Hp1-1 (Hp dimer; two Hb dimers) (B). For comparison, 1.5 μm metHb or 1.5 μm metHb-Hp1-1 mixed 1:1 with PBS are included. By estimating from the absorbance changes, 30.2 ± 0.5% (n = 3) of heme was transferred from metHb to IsdB in 300 s, whereas no measurable transfer to metHb-Hp1-1 was seen. C, relative plateau response of immobilized CD163 binding of Hb-Hp1-1 (○) and (●) Hb-Hp2-2 plotted against the indicated molar ratios of IsdBN1N2. D, sensorgrams of 100 nm Hb-Hp1-1, Hb-Hp2-2, Hb, Hp1-1, or Hp2-2 binding to immobilized IsdBN1N2. E, confocal microscopy images of endocytosis of Hb-Hp by CD163-expressing cells. CHO CD163 cells were incubated with 46 μg/ml Atto-488-labeled Hb-Hp2-2 and IsdBN1N2 in 5-fold molar excess over Hb-Hp. Atto-488-labeled Hb-Hp2-2 is shown in green, DAPI staining of nuclei is shown in blue, and CD163 surface immunostaining (Alexa 647) is shown in red. Mock transfected CHO cells (right panel) were included as a negative control.
Discussion
This study shows that S. aureus IsdH blocks the CD163-mediated uptake of Hb-Hp complexes, which instantly form when Hb is released during intravascular hemolysis. The blockage is biologically meaningful because it inhibits the normal mechanism for Hb degradation in the host and therefore secures a pool of Hb-iron for the pathogen.
The present heme transfer kinetic studies shows that IsdH is able to extract heme from metHb-Hp but not from oxyHb-Hp. This is in line with previous studies on free Hb showing that the third NEAT domain, IsdHN3, binds ferric heme tighter than ferrous heme (25) and studies with the other S. aureus receptor, IsdB, which does not extract heme from oxyHb (26). It is tempting to speculate that the IsdH blocking of the CD163-mediated pathway for Hb-Hp uptake may benefit S. aureus by providing time for the spontaneous oxidation of Hb from its ferrous form to ferric metHb, from which IsdH then can extract the heme.
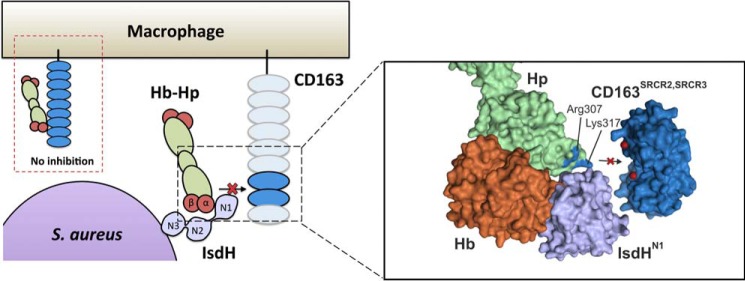
Our data indicate that both the IsdHN1 and IsdHN2 domains contribute to the binding of the Hb-Hp complexes. This is in line with previous data showing that IsdHN1 only binds the Hb α-subunit, whereas IsdHN2 binds either the Hb α- or the Hb β-subunit (27). Given that Hb-Hp complexes contain Hb αβ dimers, the higher affinity of IsdHN1N2N3 suggests that IsdHN1 and IsdHN2 are bound simultaneously to the Hb α- and β-subunits, thereby stabilizing the interaction. By presenting the known Hb-Hp-IsdHN1 crystal structure in a view together with a model of two CD163 scavenger receptor cysteine-rich (SRCR) domains, previously shown to bind Hp in the Hb-Hp complex, Fig. 9 illustrates how IsdHN1 blocks the CD163 binding site on Hb-Hp (28, 29). Fig. 9 also illustrates how two positively charged amino acids, Lys317 and Arg307, in Hp interact with acidic calcium-coordinating residues in CD163. The structural model suggests that this interaction (28) will be obstructed when IsdH binds to adjacent Hb subunit. However, x-ray crystal structures are needed to confirm the domain interactions of IsdHN1N2N3 with Hp-bound Hb.
FIGURE 9.
Model of S. aureus protein IsdH competing for CD163-mediated Hb-Hp uptake. During intravascular hemolysis, the macrophage-specific receptor CD163 normally takes up complexes of Hb-Hp. Uptake and subsequent degradation of heme produces an anti-inflammatory response. The S. aureus protein IsdH binds to Hb-Hp near the binding site of CD163, which hinders the uptake of Hb-Hp. Binding of IsdHN1 to the α-subunit of Hb in the Hb-Hp complex sterically hinders binding of Hb-Hp to CD163. The Hb-Hp-IsdHN1 structure is from Stødkilde et al. (22) (Protein Data Bank code 4WJG), whereas the two SRCR domains from CD163 were modeled from the structure of the SRCR domain of the scavenger receptor A-I (41) (Protein Data Bank code 2OY3). The positively charged residues Arg307 and Lys317 from Hp (blue) interact with negatively charged, calcium-coordinated amino acids in SRCR 2 and 3 in CD163. The calcium atoms are colored red.
IsdHN1 has, in contrast to our findings, been reported to bind Hp with a KD in the micromolar (12) or nanomolar range (13). In our study, we could not detect Hp binding by any of the IsdH constructs using either the sensitive SPR analyses or SEC-RALS in line with size exclusion chromatography. Furthermore, a recently published structure of IsdHN1 binding to Hb-Hp shows that IsdHN1 molecules only bound to Hb α-subunits (22) with no contact to Hp. Given that Hb-Hp is one of the strongest known non-covalent interactions (30), it is possible that trace contamination of Hp preparations with Hb gave rise to false positive interactions with IsdH observed in previous studies.
IsdH shares a high degree of homology with IsdB (11) and can extract heme from metHb (26). However, our data revealed that IsdB cannot extract heme from metHb-Hp, and neither is IsdB a strong inhibitor of the Hb-Hp-CD163 interaction. This clearly differentiates the role of IsdH and IsdB. Because Hb is complexed instantly with Hp in the human circulation as long as Hp exceeds the Hb concentration in plasma, the ability to sequester this pool of iron through the IsdH receptor may have provided a selective advantage during S. aureus evolution, explaining the presence of two different Hb receptors in this organism.
From a clinical point of view, IsdH blocking of Hb-Hp uptake by CD163 may worsen hemolytic sepsis by stimulating bacterial growth. Furthermore, it may enhance the inflammatory response, not only because the iron supplementation stimulates bacterial growth but also because of inhibition of Hb uptake in plasma, which is a prerequisite for conversion of heme to anti-inflammatory heme metabolites (CO and bilirubin) (31). The inhibitory effects of the S. aureus heme extraction system on the CD163-mediated Hb removal may also affect the validity of plasma Hp as a biomarker of hemolysis during S. aureus sepsis, because the well known negative correlation between Hp and hemolysis relies on Hb-Hp scavenging (32). Direct inhibition by IsdH and thereby inefficient Hp scavenging may skew this correlation and underestimate hemolysis. Finally, it is tempting to speculate that other hemolytic bacteria that extract heme from Hb might also have mechanisms to block clearance of Hb-Hp complexes.
Experimental Procedures
Protein Production
The DNA sequence encoding IsdHN1 (residues 86–229) with an added N-terminal His6 tag and a thrombin cleavage site was cloned into the NdeI and BamHI sites (Genscript) of the pET-22b(+) (Novagen) vector as previously described (22). The sequence encoding IsdHN2N3 (residues 321–655) was cloned in the XhoI and BamHI sites of pET-15b (Novagen) in line with the vector encoded His6 tag and purified over nickel affinity, anion exchange chromatography as previously described (10). IsdHN1N2N3 (residues 82–655) and IsdBN1N2 (residues 120–459) were expressed and purified according to the same method as used for IsdHN2N3 purification. The protein purity was assessed by SDS gel electrophoresis
Cell Culture
Human CD163-expressing Chinese hamster ovary cells (CHO CD163) were generated and cultured as previously described (33). Human mononuclear leukocytes were purified from blood from healthy volunteers by Ficoll-Paque centrifugation and seeded in 24-well plates (Nunc, Roskilde, Denmark) at a density of 5 × 105 monocytes/well. Monocytes were isolated by plastic adherence, as described (34) washing away non-adherent cells after 3 h of incubation at 37 °C and 5% CO2 in RPMI. The monocytes were cultured for 3 days in RPMI supplemented with 2% AB serum (Biowest, Nuaillé, France), 20 ng/ml M-CSF (Gibco, Life Technologies), and 2.5 × 10−7 m dexamethasone (Sigma-Aldrich) to increase CD163 expression as previously reported (34).
Fluorophore Labeling of Hp
Human Hp2-2 (Sigma-Aldrich) was fluorescently labeled as described previously (35). Briefly, Hp2-2 was labeled with Atto-488 succinimidyl ester (Sigma-Aldrich) by adding 65 μg of dye/mg of protein to yield a degree of labeling of 3 as confirmed by spectroscopy.
Confocal Microscopy
Confocal imaging was done as described earlier (36). Briefly, CHO CD163 or mock transfected CHO cells were seeded 3 × 104 cells/well on poly-d-lysine-coated 8-well chambered coverslips (Nunc) and cultured in serum-free CHO medium (CCM5; HyClone, Logan, UT) until the next day. Hb-Hp complexes were formed by incubating equimolar amounts of ferrous stabilized HbA0 (Sigma-Aldrich) with Atto-488-labeled Hp2-2 for 1 h. The cells were incubated for 1 h with labeled Hb-Hp2-2 (46 μg/ml) in the presence of 5-fold molar excess of IsdHN1, IsdHN2N3, or IsdHN1N2N3 (a 1:1 stoichiometry of IsdH binding to a Hb-Hp monomer unit was assumed). The cells were washed in PBS and fixed in 4% formaldehyde for 10 min at room temperature. Immunostaining of CD163 was performed by incubating the cells with 10 μg/ml rabbit anti-human CD163 IgG (Dako, Glostrup, Denmark) in 1% BSA in PBS for 45 min, followed by washing and 45 min of incubation with donkey anti-rabbit-conjugated Alexa Fluor 647 (Molecular Probes, Eugene, OR) in 1% BSA in PBS. The nuclei were stained with 300 ng/ml DAPI (Molecular Probes). The cells were visualized using an Olympus FV10i confocal microscope (Olympus Denmark A/S, Ballerup, Denmark) with a 60× NA 1.2 phase contrast water immersion objective.
Flow Cytometry
CHO cells were seeded in 24-well plates at a density of 2 × 105 cells/well. The following day the cells were incubated with Atto-488-labeled Hb-Hp2-2 (23 μg/ml) mixed with increasing ratios of IsdHN1, IsdHN2N3, or IsdHN1N2N3 in CCM5 medium for 1 h at 37 °C 5% CO2. The cells were detached by trypsination and washed twice in PBS 1% BSA. Similar experiments were performed on 72-h cultured monocytes with modifications. Briefly, uptake was performed in RPMI 1% BSA, and monocytes were detached using macrophage detachment solution (Promocell, Heidelberg, Germany) according to the manufacturer's instructions. Polyclonal rabbit anti-human CD163 IgG (Dako), earlier shown to specifically inhibit Hb-Hp uptake by CD163 (37), and irrelevant polyclonal rabbit anti-human IgG (Dako) were included as inhibition controls, both at a concentration of 50 μg/ml. Cellular uptake of fluorescent Hb-Hp2-2 and CD163 expression was analyzed by flow cytometry using a LSR II flow cytometer and FACSDiva software (BD Biosciences, San Jose, CA) using the 488- and 633-nm lasers. Geometric MFI of a population of 10,000 cells was calculated with FlowLogic software (Inivai Technologies) and used to quantify Hb-Hp uptake.
Surface Plasmon Resonance
CD163 binding to Hb-Hp with or without IsdH and IsdBN1N2 was studied by SPR on a Biacore 3000 instrument (Biacore, Uppsala, Sweden) with immobilized CD163 on a CM5 chip as previously described (29). Hb-Hp2-2 and Hb-Hp1-1 complexes were formed as described above using unlabeled Hb and diluted to 12.5 μg/ml in running buffer (10 mm HEPES, 150 mm NaCl, 2 mm CaCl2, and 0.005% Surfactant P-20, pH 7.4). The Hb-Hp complexes were mixed with increasing ratios of IsdHN1, IsdHN2N3, IsdHN1N2N3, or IsdBN1N2. Each sample (40 μl) was injected with a flow rate of 5 μl/min.
To confirm the affinity of IsdBN1N2 for Hb, Hb-Hp1-1, and Hb-Hp2-2, IsdBN1N2 was immobilized on a CM5 chip by injecting 10 μg/ml IsdBN1N2 in 10 mm sodium acetate, pH 4.0, to a surface density of ∼0.075 pmol/mm2 and capping with 1 m ethanolamine, pH 8.5. Samples (40 μl) of Hb, Hp, or Hb-Hp complexes in running buffer were injected over the chip at a flow rate of 5 μl/min in concentrations ranging from 50 to 500 nm.
For the affinity measurements of IsdH, Hp1-1 and Hp2-2 were immobilized on a CM5 chip in different flow cells in a 10 mm sodium acetate pH 4.0 buffer. Hb-Hp complexes were formed by injecting 40 μl of 100 μg/ml Hb in running buffer over the chip in each cycle followed by injection of 40 μl of 25, 50, 100, 200, 300, 400, or 500 nm IsdHN1, IsdHN2N3, or IsdHN1N2N3 at a flow rate of 5 μl/min. Regeneration was obtained by injecting two cycles of 10 μl of regeneration buffer (10 mm glycin, 20 mm EDTA, 500 mm NaCl, and 0.005% Surfactant P-20, pH 3.0). In parallel, endoglycosidase H, expressed with a His6 tag, was used as a negative control to exclude the possibility of the His tag binding to Hb-Hp or Hp. The data were analyzed using BIAevaluation software 4.0.1, and the apparent dissociation constants were obtained by fitting the binding response of IsdH constructs to Hb-Hp1-1 and Hb-Hp2-2 flow cells just prior to the end of injection (RPl) to the previously described expression (38).
| (Eq. 1) |
Heme Transfer Kinetics
The transfer rate of heme from free Hb and from Hp1-1-bound Hb to IsdH was measured using a Cary 60 (Agilent Technologies, Glostrup, Denmark) UV-visible spectrophotometer at 20 °C. Pure ferrous adult human HbA0 (Sigma-Aldrich) in the oxygenated form (oxyHb) was oxidized to the ferric (met) form (metHb) by ferricyanide, as described (39). Briefly, the lyophilized Hb was dissolved in milliQ water at a concentration of 4–6 mm heme and incubated in the dark at room temperature for 1 h with a 1.2 molar excess of potassium ferricyanide K3Fe(CN)6 over heme. MetHb was desalted on a PD-10 column (GE Healthcare) equilibrated with 100 mm HEPES, pH 7.5, to remove ferricyanide and dialyzed against milliQ water before use.
Concentrations of metHb and oxyHb were determined using extinction coefficients 179 mm−1 cm−1 at 405 nm and 14.6 mm−1 cm−1 at 577 nm, respectively (39). Hb-Hp complexes were made by mixing 1.5 μm (tetrameric) metHb or oxyHb with a 1.5 molar excess of Hp1-1 (Sigma-Aldrich) in PBS (pH 7.4), assuming that one Hp1-1 dimer binds two Hb dimers. IsdHN2N3, IsdHN1, or IsdBN1N2 (all 13 μm in PBS) was mixed in a 1-cm quartz cuvette 1:1 with either metHb or oxyHb (both 1.5 μm in PBS) alone or bound to Hp1-1. Absorbance spectra were recorded every 5 s in the range 350–450 nm for 300 s.
Size Exclusion Chromatography with In-line Right Angle Light Scattering
SEC was performed on a Viscotek P2500 column (Malvern, Worcestershire, UK). RALS and refractive index were measured on a Viscotek 305 Triple Detector Array instrument (Malvern). The detectors and column were maintained at 30 °C. The light scattering cell was illuminated by a laser diode (670 nm), and light scattered at an angle of 90° was measured by a photodiode detector. The refractive index detector was a dual cell design. Calibration of the detectors and calculation of sample weight-average molecular mass was performed using the Omnisec software (Malvern). The instrument was calibrated using multiple protein and polyethylene oxide standards. Sample molecular mass was calculated using a specific refractive index increment with respect to sample concentration (dn/dc) of 0.185 g/ml, neglecting possible deviations because of the presence of co-factors or glycosylation.
Author Contributions
K. L. S. performed experiments and manuscript writing; K. S. performed experiments and manuscript writing; J. H. G. performed experiments (Biacore); C. F. D. performed experiments (SEC-RALS and recombinant protein purification); D. G. performed experiments (SEC-RALS and recombinant protein purification); A. E. performed experiments (gene transfection); C. B. F. A. performed experiments (structure analysis); S. W. K. H. performed experiments (supervision of flow cytometry); A. F. performed experiments (supervision of heme-transfer kinetics); and S. K. M. performed study design, supervision, and manuscript writing.
Acknowledgments
We thank Anne-Marie Bundsgaard and Patrick Bjork Richardt for technical assistance.
This work was supported by the Novo Nordisk Foundation, the Lundbeck Foundation, and Danish Medical Research Council and European Research Council TROJA Grant 233312). The authors declare that they have no conflicts of interest with the contents of this article.
- Isd
- iron-regulated surface determinant
- NEAT
- near iron transporter
- hb
- hemoglobin
- Hp
- haptoglobin
- SEC
- size exclusion chromatography
- RALS
- in-line right angle light scattering
- CHO CD163
- CD163-expressing CHO cells
- SRCR
- scavenger receptor cysteine-rich
- MFI
- mean fluorescence intensity
- SPR
- surface plasmon resonance.
References
- 1. Gorwitz R. J., Kruszon-Moran D., McAllister S. K., McQuillan G., McDougal L. K., Fosheim G. E., Jensen B. J., Killgore G., Tenover F. C., and Kuehnert M. J. (2008) Changes in the prevalence of nasal colonization with Staphylococcus aureus in the United States, 2001–2004. J. Infect. Dis. 197, 1226–1234 [DOI] [PubMed] [Google Scholar]
- 2. Eichenbaum Z., Muller E., Morse S. A., and Scott J. R. (1996) Acquisition of iron from host proteins by the group A streptococcus. Infect. Immun. 64, 5428–5429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Johnson M. K., and Boese-Marrazzo D. (1980) Production and properties of heat-stable extracellular hemolysin from Pseudomonas aeruginosa. Infect. Immun. 29, 1028–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Smith H. W. (1963) The haemolysins of Escherichia coli. J. Pathol. Bacteriol. 85, 197–211 [DOI] [PubMed] [Google Scholar]
- 5. Mazmanian S. K., Skaar E. P., Gaspar A. H., Humayun M., Gornicki P., Jelenska J., Joachmiak A., Missiakas D. M., and Schneewind O. (2003) Passage of heme-iron across the envelope of Staphylococcus aureus. Science 299, 906–909 [DOI] [PubMed] [Google Scholar]
- 6. Mazmanian S. K., Ton-That H., Su K., and Schneewind O. (2002) An iron-regulated sortase anchors a class of surface protein during Staphylococcus aureus pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 99, 2293–2298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sheldon J. R., and Heinrichs D. E. (2015) Recent developments in understanding the iron acquisition strategies of gram positive pathogens. FEMS Microbiol. Rev. 39, 592–630 [DOI] [PubMed] [Google Scholar]
- 8. Fonner B. A., Tripet B. P., Eilers B. J., Stanisich J., Sullivan-Springhetti R. K., Moore R., Liu M., Lei B., and Copié V. (2014) Solution structure and molecular determinants of hemoglobin binding of the first NEAT domain of IsdB in Staphylococcus aureus. Biochemistry 53, 3922–3933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Spirig T., Malmirchegini G. R., Zhang J., Robson S. A., Sjodt M., Liu M., Krishna Kumar K., Dickson C. F., Gell D. A., Lei B., Loo J. A., and Clubb R. T. (2013) Staphylococcus aureus uses a novel multidomain receptor to break apart human hemoglobin and steal its heme. J. Biol. Chem. 288, 1065–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dickson C. F., Kumar K. K., Jacques D. A., Malmirchegini G. R., Spirig T., Mackay J. P., Clubb R. T., Guss J. M., and Gell D. A. (2014) Structure of the hemoglobin-IsdH complex reveals the molecular basis of iron capture by Staphylococcus aureus. J. Biol. Chem. 289, 6728–6738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sjodt M., Macdonald R., Spirig T., Chan A. H., Dickson C. F., Fabian M., Olson J. S., Gell D. A., and Clubb R. T. (2016) The PRE-derived NMR model of the 38.8-kDa tri-domain IsdH protein from Staphylococcus aureus suggests that it adaptively recognizes human hemoglobin. J. Mol. Biol. 428, 1107–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dryla A., Hoffmann B., Gelbmann D., Giefing C., Hanner M., Meinke A., Anderson A. S., Koppensteiner W., Konrat R., von Gabain A., and Nagy E. (2007) High-affinity binding of the staphylococcal HarA protein to haptoglobin and hemoglobin involves a domain with an antiparallel eight-stranded beta-barrel fold. J. Bacteriol. 189, 254–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pilpa R. M., Robson S. A., Villareal V. A., Wong M. L., Phillips M., and Clubb R. T. (2009) Functionally distinct NEAT (NEAr Transporter) domains within the Staphylococcus aureus IsdH/HarA protein extract heme from methemoglobin. J. Biol. Chem. 284, 1166–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Visai L., Yanagisawa N., Josefsson E., Tarkowski A., Pezzali I., Rooijakkers S. H., Foster T. J., and Speziale P. (2009) Immune evasion by Staphylococcus aureus conferred by iron-regulated surface determinant protein IsdH. Microbiology 155, 667–679 [DOI] [PubMed] [Google Scholar]
- 15. Alayash A. I., Andersen C. B., Moestrup S. K., and Bülow L. (2013) Haptoglobin: the hemoglobin detoxifier in plasma. Trends Biotechnol. 31, 2–3 [DOI] [PubMed] [Google Scholar]
- 16. Andersen C. B., Torvund-Jensen M., Nielsen M. J., de Oliveira C. L., Hersleth H. P., Andersen N. H., Pedersen J. S., Andersen G. R., and Moestrup S. K. (2012) Structure of the haptoglobin-haemoglobin complex. Nature 489, 456–459 [DOI] [PubMed] [Google Scholar]
- 17. Andersen C. B., Stødkilde K., Sæderup K. L., Kuhlee A., Raunser S., Graversen J. H., and Moestrup S. K. (2016) Haptoglobin. Antioxidant Redox Signal., in press [DOI] [PubMed] [Google Scholar]
- 18. Buehler P. W., Abraham B., Vallelian F., Linnemayr C., Pereira C. P., Cipollo J. F., Jia Y., Mikolajczyk M., Boretti F. S., Schoedon G., Alayash A. I., and Schaer D. J. (2009) Haptoglobin preserves the CD163 hemoglobin scavenger pathway by shielding hemoglobin from peroxidative modification. Blood 113, 2578–2586 [DOI] [PubMed] [Google Scholar]
- 19. Kristiansen M., Graversen J. H., Jacobsen C., Sonne O., Hoffman H. J., Law S. K., and Moestrup S. K. (2001) Identification of the haemoglobin scavenger receptor. Nature 409, 198–201 [DOI] [PubMed] [Google Scholar]
- 20. Lim S. K., Kim H., Lim S. K., bin Ali A., Lim Y. K., Wang Y., Chong S. M., Costantini F., and Baumman H. (1998) Increased susceptibility in Hp knockout mice during acute hemolysis. Blood 92, 1870–1877 [PubMed] [Google Scholar]
- 21. Schaer D. J., Buehler P. W., Alayash A. I., Belcher J. D., and Vercellotti G. M. (2013) Hemolysis and free hemoglobin revisited: exploring hemoglobin and hemin scavengers as a novel class of therapeutic proteins. Blood 121, 1276–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stødkilde K., Torvund-Jensen M., Moestrup S. K., and Andersen C. B. (2014) Structural basis for trypanosomal haem acquisition and susceptibility to the host innate immune system. Nat. Commun. 5, 5487. [DOI] [PubMed] [Google Scholar]
- 23. Dryla A., Gelbmann D., von Gabain A., and Nagy E. (2003) Identification of a novel iron regulated staphylococcal surface protein with haptoglobin-haemoglobin binding activity. Mol. Microbiol. 49, 37–53 [DOI] [PubMed] [Google Scholar]
- 24. Valdes R. Jr., Vickers L. P., Halvorson H. R., and Ackers G. K. (1978) Reciprocal effects in human hemoglobin: direct measurement of the dimer-tetramer association constant at partial oxygen saturation. Proc. Natl. Acad. Sci. U.S.A. 75, 5493–5496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Moriwaki Y., Caaveiro J. M., Tanaka Y., Tsutsumi H., Hamachi I., and Tsumoto K. (2011) Molecular basis of recognition of antibacterial porphyrins by heme-transporter IsdH-NEAT3 of Staphylococcus aureus. Biochemistry 50, 7311–7320 [DOI] [PubMed] [Google Scholar]
- 26. Bowden C. F., Verstraete M. M., Eltis L. D., and Murphy M. E. (2014) Hemoglobin binding and catalytic heme extraction by IsdB near iron transporter domains. Biochemistry 53, 2286–2294 [DOI] [PubMed] [Google Scholar]
- 27. Krishna Kumar K., Jacques D. A., Pishchany G., Caradoc-Davies T., Spirig T., Malmirchegini G. R., Langley D. B., Dickson C. F., Mackay J. P., Clubb R. T., Skaar E. P., Guss J. M., and Gell D. A. (2011) Structural basis for hemoglobin capture by Staphylococcus aureus cell-surface protein, IsdH. J. Biol. Chem. 286, 38439–38447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nielsen M. J., Andersen C. B., and Moestrup S. K. (2013) CD163 binding to haptoglobin-hemoglobin complexes involves a dual-point electrostatic receptor-ligand pairing. J. Biol. Chem. 288, 18834–18841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Madsen M., Møller H. J., Nielsen M. J., Jacobsen C., Graversen J. H., van den Berg T., and Moestrup S. K. (2004) Molecular characterization of the haptoglobin.hemoglobin receptor CD163. Ligand binding properties of the scavenger receptor cysteine-rich domain region. J. Biol. Chem. 279, 51561–51567 [DOI] [PubMed] [Google Scholar]
- 30. Hwang P. K., and Greer J. (1980) Interaction between hemoglobin subunits in the hemoglobin. haptoglobin complex. J. Biol. Chem. 255, 3038–3041 [PubMed] [Google Scholar]
- 31. Etzerodt A., and Moestrup S. K. (2013) CD163 and inflammation: biological, diagnostic, and therapeutic aspects. Antioxid. Redox Signal. 18, 2352–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Muller-Eberhard U., Javid J., Liem H. H., Hanstein A., and Hanna M. (1968) Plasma concentrations of hemopexin, haptoglobin and heme in patients with various hemolytic diseases. Blood 32, 811–815 [PubMed] [Google Scholar]
- 33. Nielsen M. J., Madsen M., Møller H. J., and Moestrup S. K. (2006) The macrophage scavenger receptor CD163: endocytic properties of cytoplasmic tail variants. J. Leukoc. Biol. 79, 837–845 [DOI] [PubMed] [Google Scholar]
- 34. Schaer D. J., Boretti F. S., Schoedon G., and Schaffner A. (2002) Induction of the CD163-dependent haemoglobin uptake by macrophages as a novel anti-inflammatory action of glucocorticoids. Br. J. Haematol. 119, 239–243 [DOI] [PubMed] [Google Scholar]
- 35. Boretti F. S., Baek J. H., Palmer A. F., Schaer D. J., and Buehler P. W. (2014) Modeling hemoglobin and hemoglobin:haptoglobin complex clearance in a non-rodent species-pharmacokinetic and therapeutic implications. Front. Physiol. 5, 385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Etzerodt A., Maniecki M. B., Graversen J. H., Møller H. J., Torchilin V. P., and Moestrup S. K. (2012) Efficient intracellular drug-targeting of macrophages using stealth liposomes directed to the hemoglobin scavenger receptor CD163. J. Control Release 160, 72–80 [DOI] [PubMed] [Google Scholar]
- 37. Schaer C. A., Vallelian F., Imhof A., Schoedon G., and Schaer D. J. (2007) CD163-expressing monocytes constitute an endotoxin-sensitive Hb clearance compartment within the vascular system. J. Leukoc. Biol. 82, 106–110 [DOI] [PubMed] [Google Scholar]
- 38. MacKenzie C. R., Hirama T., Deng S. J., Bundle D. R., Narang S. A., and Young N. M. (1996) Analysis by surface plasmon resonance of the influence of valence on the ligand binding affinity and kinetics of an anti-carbohydrate antibody. J. Biol. Chem. 271, 1527–1533 [DOI] [PubMed] [Google Scholar]
- 39. Antonini E., and Brunori M. (1971) Hemoglobin and Myoglobin in Their Reactions with Ligands 1, North-Holland Publishing Company, Amsterdam, The Netherlands [Google Scholar]
- 40. Skaar E. P., and Schneewind O. (2004) Iron-regulated surface determinants (Isd) of Staphylococcus aureus: stealing iron from heme. Microbes Infect. 6, 390–397 [DOI] [PubMed] [Google Scholar]
- 41. Ojala J. R., Pikkarainen T., Tuuttila A., Sandalova T., and Tryggvason K. (2007) Crystal structure of the cysteine-rich domain of scavenger receptor MARCO reveals the presence of a basic and an acidic cluster that both contribute to ligand recognition. J. Biol. Chem. 282, 16654–16666 [DOI] [PubMed] [Google Scholar]