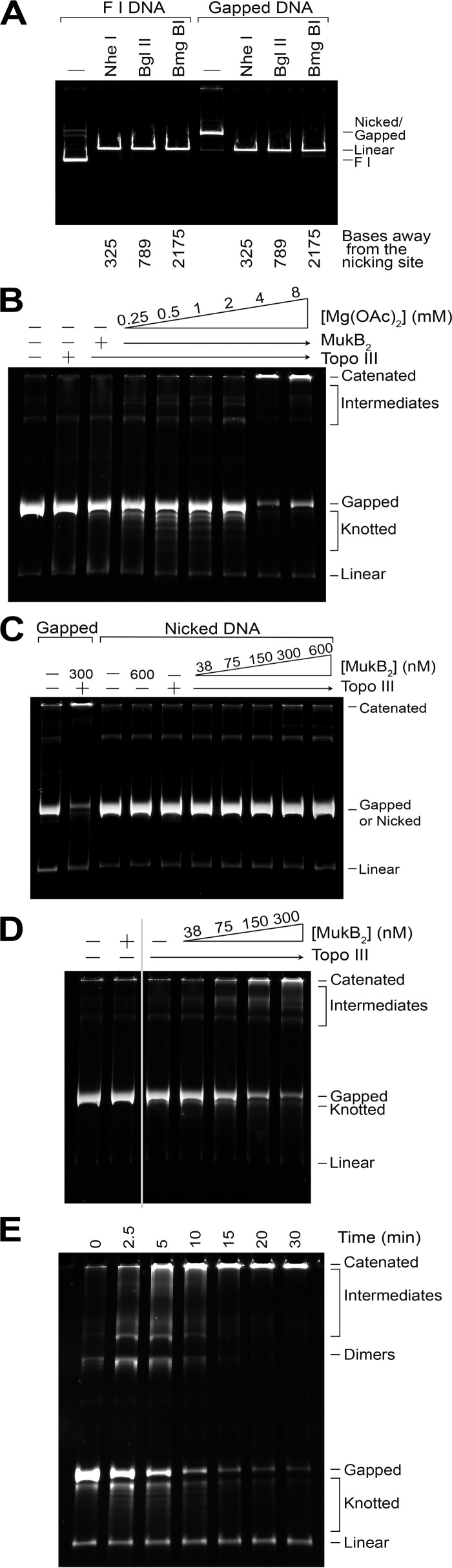
FIGURE 1.
MukB-mediated catenation of DNA. A, the gapped DNA substrate has gaps that are less than 325 nucleotides in length. Either supercoiled pCG09 form I DNA (F1 DNA) or gapped pCG09 DNA prepared as described under “Experimental Procedures,” were treated with the indicated restriction enzymes that cut the plasmid DNA only once at the distance indicated from the nicking site. Complete digestion to linear product indicates that the DNA must be double-stranded at the indicated site. B, in the presence of Topo III, MukB mediates knotting of single DNA rings at low Mg2+ concentrations, but mediates catenation of DNA rings at moderate Mg2+ concentrations. Reaction mixtures containing 0.7 nm gapped DNA, 25 nm Topo III (a plus above the lane followed by an arrow indicates that all reactions analyzed in lanes covered by the arrow contained the indicated component), 300 nm MukB (as dimer), as indicated, and the indicated concentrations of Mg(OAc)2, were incubated for 30 min at 37 °C, processed, and analyzed as described under “Experimental Procedures.” C, a gap is required to observe DNA catenation. Standard reaction mixtures containing either gapped DNA or DNA that had only been nicked with Nb.BbVCI (nicked DNA), 25 nm Topo III, as indicated, and the indicated concentration of MukB were incubated for 30 min at 37 °C, processed, and analyzed as described under “Experimental Procedures.” D, MukB-mediated catenation of DNA in the presence of Topo III as a function of MukB concentration. Standard reaction mixtures containing 25 nm Topo III and the indicated concentrations of MukB were incubated for 5 min at 37 °C, processed, and analyzed as described under “Experimental Procedures.” E, time course of DNA catenation at 300 nm MukB. A standard reaction mixture was increased 8-fold in size in volume and incubated at 37 °C. Aliquots (20 μl) were removed at the indicated times and processed and analyzed as described under “Experimental Procedures.” Note that the zero time aliquot was removed after mixing all components of the reaction together, accounting for the slight catenation and knotting observed. Catenated, large networks of catenated DNA rings that do not enter the gel, some of this material will wash off the top of the gels during handling. Intermediates, less complicated catenated networks of DNA rings were probably made of chains of DNA rings linked once. Gapped, gDNA starting material. Knotted, single DNA rings that have become knotted. Linear, a trace amount of linear pCG09 DNA present in the gDNA preparation. All assays were repeated at least three times, representative gels are shown.