FIGURE 4.
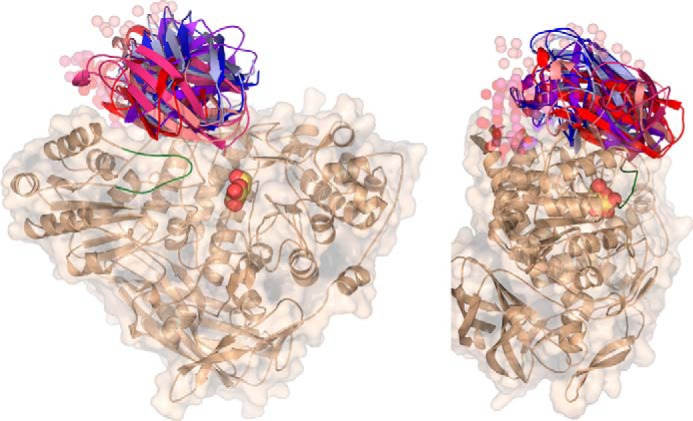
SAXS analysis of GlyA1. Six ab initio models were generated for complete GlyA1 from SAXS data, using the experimental structure of the truncated protein and two different models of the last 120 residues (GlyA1-Ct). The two templates were obtained from Swiss-Model (red) (48) or CPHmodel (blue) (49) servers, which predict different lengths of the linker attaching this domain to the core protein, 32 or 5 residues, respectively. CORAL (47) modeling of this linker in each run is represented in spheres. The active site pocket is indicated by the galactose found at the crystal (yellow), and the mobile loop (residues 100–113), as observed in the galactose-soaked crystals, is highlighted in green.
