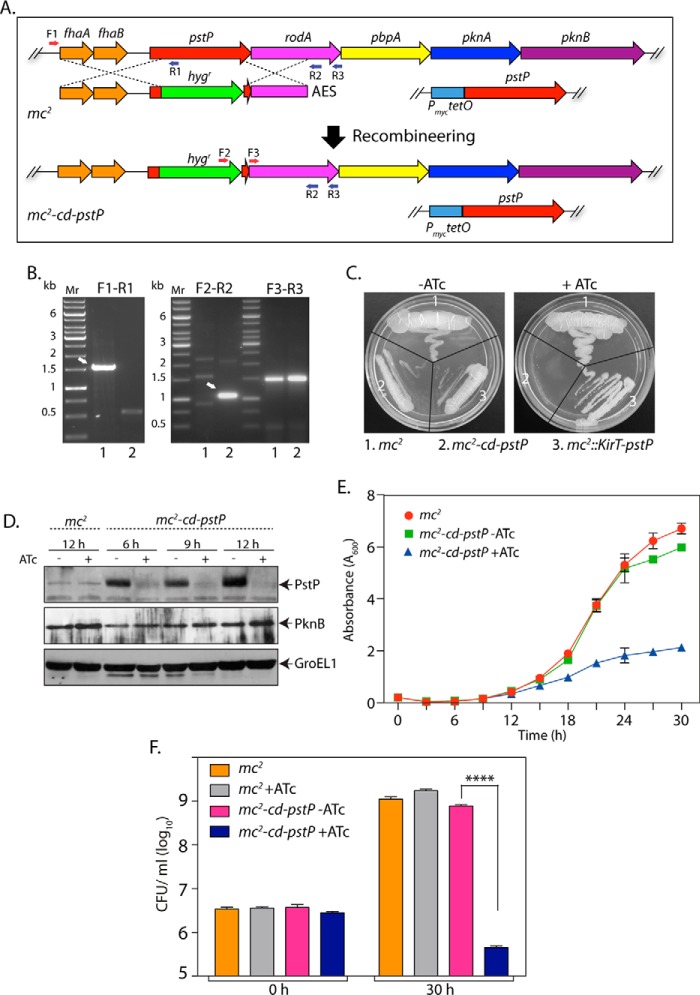
FIGURE 3.
Depletion of PstP in M. smegmatis leads to poor cell survival. A, schematic depiction of generation of gene replacement mutant using the recombineering method. Primers used for screening the mutant are shown. B, agarose gels showing PCR products obtained using different primer pairs with genomic DNA obtained from mc2 (lane 1) and putative mc2-cd-pstP mutant (lane 2). F3-R3 amplifying a region of the rodA gene (∼1.5 kb) was used as a control. F1-R1, ∼1.5-kb fragment for mc2 and no amplicon expected for mc2-cd-pstP. F2-R2, no amplicon expected for mc2 and ∼1.1 kb for mc2-cd-pstP. C, mc2, mc2::pKirT-pstP, and mc2-cd-pstP cultures were grown until A600 reached ∼0.6, and the cultures were diluted to an A600 of 0.1. Diluted cultures were streaked on 7H10 agar plates not containing or containing 50 ng/ml ATc. D, mc2 and mc2-cd-pstP cultures were initiated at an A600 of 0.1 and grown in the absence or presence of ATc for 6, 9, or 12 h. Whole cell lysates were isolated, resolved on SDS-PAGE, transferred to nitrocellulose membrane, and probed with α-PstP, α-PknB, and α-GroEL1 antibodies. E, mc2 and mc2-cd-pstP cultures were initiated at an initial A600 of 0.02 and grown in the absence or presence of ATc for 30 h. The experiment was performed in triplicates, and the error bar represents S.E. F, cfu were enumerated at 0 and 30 h for both mc2 and mc2-cd-pstP in the presence and absence of ATc. cfu data were analyzed and plotted as mean with S.D. (error bars) using GraphPad Prism version 6, and significance was calculated using two-way ANOVA with p < 0.0001 (****).