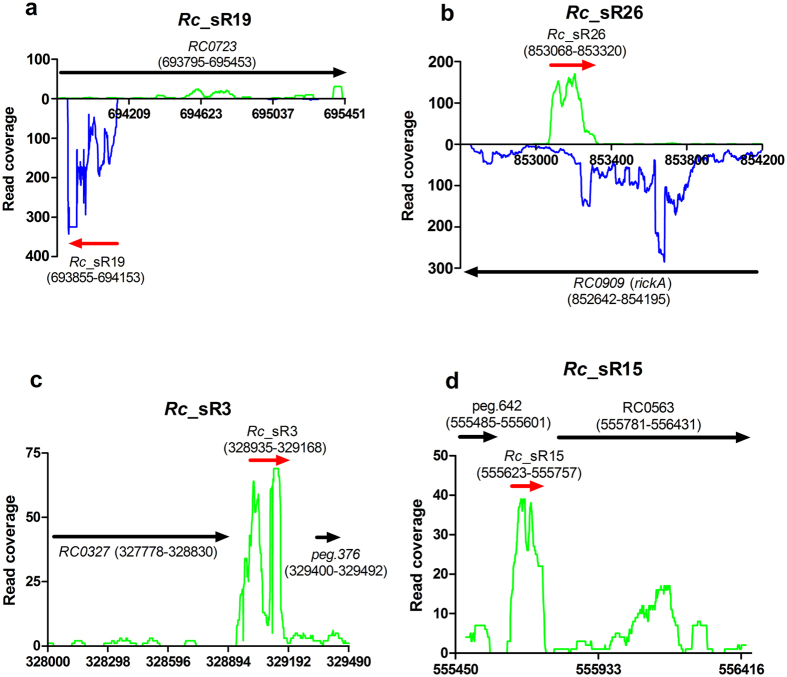
Figure 2. Representative graphs showing the read coverage of novel sRNAs in R. conorii during the infection of HMECs.
HMECs were infected with R. conorii (MOI = 20) and total RNA was harvested at 3 and 24 h post-infection. High throughput RNA sequencing was performed on total RNA enriched for bacterial transcripts (see materials and methods). The strand specific reads were mapped onto R. conorii genome (PATRIC genome ID: 27944.4). The read coverage plots of two cis-acting sRNAs (a) Rc_sR19 and (b) Rc_sR26, and two riboswitches (c) Rc_sR3 and (d) Rc_sR15 are presented. The reads above and below the x-axis represent the forward (green) and reverse strand (blue), respectively. The identified sRNAs are indicated by red arrow. The ORFs up- and downstream, and cis-acting ORF are shown by black arrow. The orientation and genomic location coordinates correspond to the R. conorii genome annotation in PATRIC.