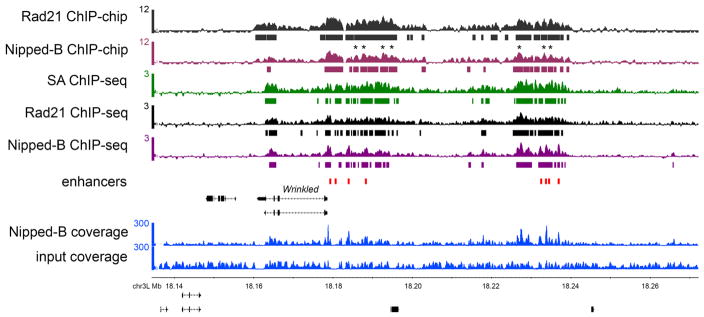
Figure 1.
ChIP-chip and ChIP-seq of cohesin and Nipped-B. The browser view show the Wrinkled gene and its predicted transcriptional enhancers (red boxes) (8). The ChIP-chip track scales are MAT scores (10) and the bars underneath indicate where enrichment is significant at a p value ≤ 0.001. The ChIP-seq track scales are log2 enrichment (ChIP/input), and the bars underneath indicate where enrichment is ≥ 2σ for ≥ 300 bp (six windows), which gives a similar pattern as the statistical threshold used for ChIP-chip. The ChIP sample and input coverage tracks used to calculate the Nipped-B enrichments are shown. The ChIP-chip and ChIP-seq tracks show very similar global enrichment patterns, but for both Rad21 and Nipped-B there are cases where the ChIP-chip peaks are valleys in the ChIP-seq track. Some of these are marked with asterisks (*) above the Nipped-B ChIP-chip track. These are usually where input and ChIP sequence coverage are lower than average, suggesting that normalization of ChIP to input incompletely compensates for regions that amplify or sequence poorly.